Figure 4.
90A RNP Recognition by the Pan2 RNase Domain and the Pan3straight Pseudokinase Domain
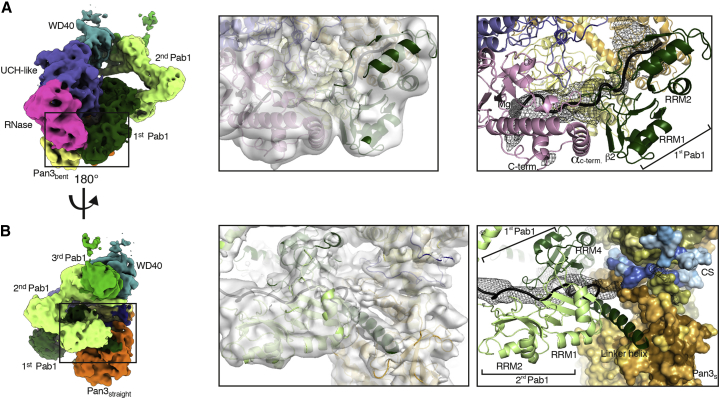
(A) Views of the interaction between UCH-like-RNase modules of Pan2 (violet and pink, respectively) and the RRM1-RRM2 module of the first Pab1 (dark green). The pseudo-atomic model (right panel) is superposed on the cryo-EM density (central panel). The left panel identifies the overall position of the interface in the context of the reconstruction (shown as segmented density, as in Figure 3B). Difference density for the RNA is shown in mesh representation, in black. The directionality of RNase-Pab1 recognition is fixed by the defined polarity of the poly(A) recognition by RRM1-RRM2 (3′ end at the N terminus, 5′ end at the C terminus) as well as the 3′-to-5′ exonuclease activity of Pan2 (additional details in Figure S5A).
(B) Corresponding views of the interaction between the Pan3s pseudokinase domain and the first RNP oligomerization interface (i.e., RRM4-linker helix of the first Pab1 protomer and the RRM1-RRM2 module of the second protomer). Pan3s is shown in a surface representation colored according to evolutionary conservation (dark orange for conserved residues). The RNP contacts the connecting segment (CS) of Pan2, also shown in a surface representation and colored according to conservation (conserved residues in dark blue; see also Figure S5B).