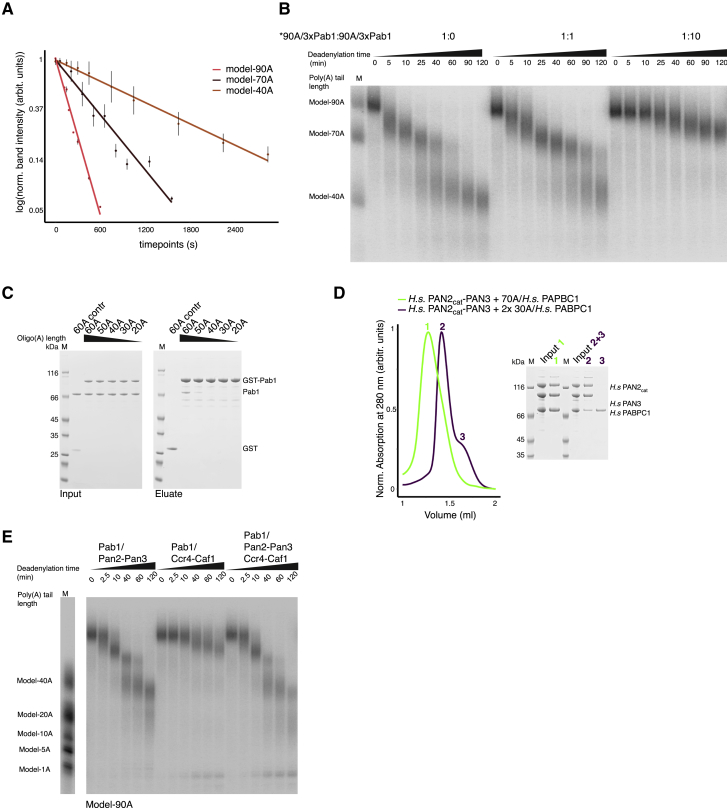
Figure S1.
Yeast and Human Pan2-Pan3 Complexes Preferentially Bind and Deadenylate poly(A) RNPs, Related to Figure 2
(A) Pan2-Pan3 is more active on longer poly(A)/Pab1 RNPs in in vitro deadenylation experiments. Three phosphorimages of UREA-PAGE gels of Pan2-Pan3 mediated deadenylation assays with a model-90A RNP as educt (similar and including Figure 1C) were quantified by densitometry. After normalization and natural log-transformation, median band intensities per time point for the model-90A RNP as well as the two intermediates, the model-70A RNP and the model-40A RNP are plotted. In all three cases the respective timepoint 0 corresponds to the timepoint in the assay with the most intense densitometric reading for the respective model substrate. Vertical lines represent the standard deviation interval at each particular time-point. The trend lines are the curves fitted to determine the half-lives of the respective model poly(A) RNPs (see Table S2).
(B) Poly(A) tail length preference of Pan2-Pan3. The 5′-labeled 90A RNP was subjected to deadenylation in the presence of increasing amounts of unlabeled 90A RNP. The ratios between radioactive (hot) 90A and not radioactive (cold) 90A used in the competition assays are specified above each time course (each RNA substrate was incubated independently with Pab1 prior to the reaction). At indicated time points samples were taken and analyzed on a 6% Urea-PAGE followed by phosphorimaging.
(C) Interaction of multiple Pab1 protomer on longer oligo(A) RNAs. We incubated GST-tagged and untagged Pab1 in the presence of oligo(A) RNAs of different length, ranging between 20A and 60A, and tested the interaction in pull-down assays with glutathione-agarose beads. In this experiment, an increase from 40A to 60A was required to observe significant Pab1-Pab1 co-precipitation. The Coomassie-stained 4%–12% SDS-PAGE gels show the input (left) and the pulled-down protein precipitates (right).
(D) The interaction of human PAN2-PAN3 and PABPC1 depends on poly(A) tail length. Size-exclusion chromatography (SEC) assays carried out with a PAN2cat-PAN3 catalytically inactive mutant (D1087A, orthologues to D1020A in S. cerevisiae). In case of the comigration experiments of PAN2cat-PAN3 and 30A/PABPC1 we used 2x molar excess of the RNP to ensure a comparable concentration of binding sites and bases present as in the longer poly(A) RNPs. Left panel: overlays of the chromatograms. Right panel: Coomassie-stained 4%–12% SDS–PAGE gels with samples from the input and peak fractions.
(E) Deadenylation time course of a model-90A RNP (1:3 RNA:Pab1 ratio) upon addition of either Pan2-Pan3 or a Caf1-Ccr4 complex (Basquin et al., 2012), or both. The reactions were stopped at the indicated time points and analyzed on a 6% Urea-PAGE followed by phosphorimaging.