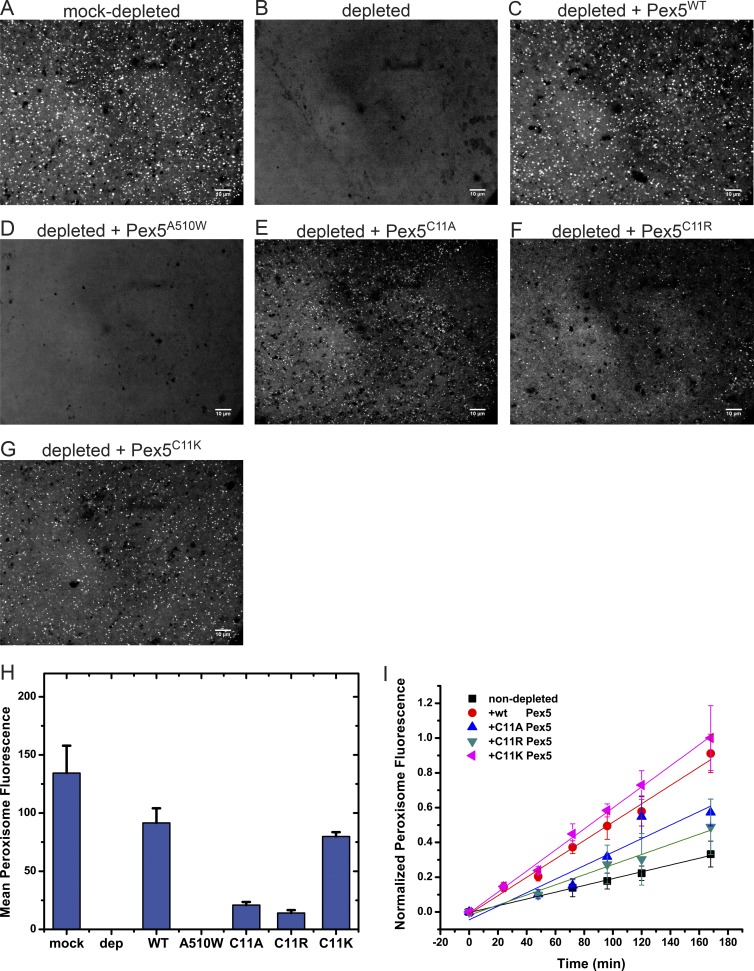
Figure 6.
Peroxisome protein import with Cys11 mutations in Pex5. (A) Xenopus cleared egg extract was mock-depleted with beads, as in Fig. 3, and incubated with 0.6 µM mScarlet-SKL for 1 h at 20–23°C. The sample was imaged with a spinning-disk confocal microscope. (B) As in A, but with an extract depleted of Pex5 with beads containing affinity-purified Pex5 antibodies (see Fig. 3). (C) As in B, but with 1 µM WT Pex5 added. (D) As in B, but with 1 µM Pex5A510W, defective in SKL binding. (E) As in B, but with 1 µM Pex5C11A. (F) As in B, but with 1 µM Pex5C11R. (G) As in B, but with 1 µM Pex5C11K. (H) Quantification of the end-point fluorescence in peroxisomes, using automated image analysis. Shown are the combined data of two parallel experiments (>20 images). The mean and standard deviation of the mean are given. (I) WT Pex5 or the indicated Cys11 mutants were added at 1 µM to cleared, un-depleted extract. The samples were imaged at the indicated time points with a spinning-disk confocal microscope, and the mean fluorescence per peroxisome was determined by automated image analysis. Shown are the combined data of two experiments done on different days (>20 images per time point), each normalized to the final time point of the brightest sample. For each time point, the mean and the standard deviation of the mean are given. Bars, 10 µm.