Figure 2. Yeast-display directed evolution to improve split APEX2.
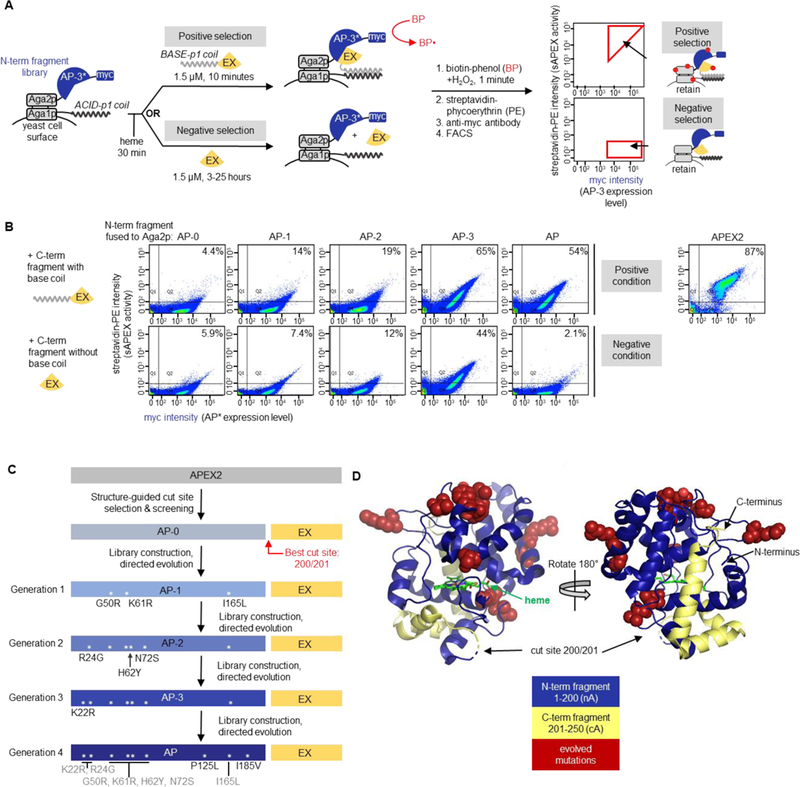
(A) Yeast display-based directed evolution scheme. The scheme shown here was used for Generation 4 selections, but the setup for other generations was similar (detailed in Figures S4–S7). A library of N-terminal fragments based on “AP-3” (the winning clone from Generation 3 selections) was displayed on the yeast cell surface via fusion to the Aga2p mating protein. An acid coil was co-displayed, via fusion to Aga1p, to recruit base coil-fused C-terminal fragment (“EX”, amino acids 201–250 of APEX2). In the positive selection for high sAPEX activity, base coil-EX-GFP was incubated with the AP-3 yeast library for 10 minutes, then reconstituted peroxidase activity was detected by treating the cell mixture with biotin-phenol (BP) and H2O2. Cells with high peroxidase activity label themselves with biotin to a high extent16, enabling their enrichment via FACS after streptavidin-phycoerythryin (PE) staining. In the negative selection to deplete the sAPEX library of AP-3 fragments with excessively high affinity for EX, we incubated the AP-3 yeast library with EX-GFP protein lacking base coil for increasingly long time periods (see Figure S7A), then performed BP labeling. Cells with low streptavidin-PE signal were retained via FACS. Myc staining on the x-axis provides a readout of AP expression level. Note that the fluorescence from GFP was not measured during FACS; GFP was included to increase the solubility of EX. Hypothetical data are shown to illustrate the strategy for gate selection. (B) Summary of improvement of sAPEX activity and PPI-dependence throughout generations of selections in yeast. Full length APEX2 fusion to Aga2p was used as a benchmark for desired activity range. Yeast were prepared as in (A), with the indicated N-terminal fragment of sAPEX expressed on the yeast surface as a fusion to Aga2p. Purified C-terminal fragment (EX), either with or without acid coil, was added to the cells for 10 minutes or 7 hours, respectively. Then BP labeling, streptavidin-PE staining, and FACS were performed as in (A). AP-1, AP-2, and AP-3 are the first, second, and third generation N-terminal fragment clones, whose mutations are shown in (C). The percentage of myc-positive cells in the top right quadrant (Q2) is indicated in the top right corner of each FACS plot. Data are shown for one out of two biological replicates. (C) Summary of split APEX protein engineering. The name of the best N-terminal fragment clone to emerge from Generation 1 selection is “AP-1”, and so on, as indicated. The best clone to emerge from the Generation 4 selection is “AP.” Asterisks depict the locations of mutations within the protein sequence. (D) sAPEX split site and mutations in sAPEX relative to full-length APEX22. The split site is between E200 and G201, and the N-terminal fragment (“AP”) and C-terminal fragment (“EX”) of sAPEX are colored blue and yellow, respectively. The nine residues mutated through directed evolution are colored red and rendered in space-filling mode; the original residues from the parent protein are depicted. Structures based on PBD ID: 1OAG55.
