Figure 1.
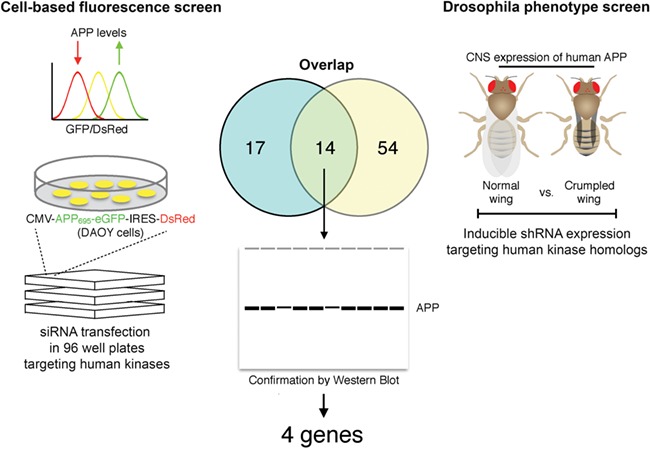
Schematic diagram of the cross-species kinome RNAi screen. Kinases capable of modifying APP stability were identified by fluorescence sorting in Daoy cells stably transfected with APP-eGFP-IRES-DsRed (left). A parallel screen was conducted in APP transgenic Drosophila using the crumpled wing phenotype as readout for neuronal APP levels (right). Candidate modifiers identified from each screen were compared to identify 14 shared genes (center). These 14 shared modifiers were tested individually in each system by Western blot to define 4 confirmed kinase modifiers of APP (center).
