An amino acid sequence motif in the N-acetylglucosaminyltransferase I transmembrane domain governs its Golgi localization and glycan processing function in plants.
Abstract
The Golgi apparatus consists of stacked cisternae filled with enzymes that facilitate the sequential and highly controlled modification of glycans from proteins that transit through the organelle. Although the glycan processing pathways have been extensively studied, the underlying mechanisms that concentrate Golgi-resident glycosyltransferases and glycosidases in distinct Golgi compartments are poorly understood. The single-pass transmembrane domain (TMD) of n-acetylglucosaminyltransferaseI (GnTI) accounts for its steady-state distribution in the cis/medial-Golgi. Here, we investigated the contribution of individual amino acid residues within the TMD of Arabidopsis (Arabidopsis thaliana) and Nicotiana tabacum GnTI toward Golgi localization and n-glycan processing. Conserved sequence motifs within the TMD were replaced with those from the established trans-Golgi enzyme α2,6-sialyltransferase and site-directed mutagenesis was used to exchange individual amino acid residues. Subsequent subcellular localization of fluorescent fusion proteins and n-glycan profiling revealed that a conserved Gln residue in the GnTI TMD is essential for its cis/medial-Golgi localization. Substitution of the crucial Gln residue with other amino acids resulted in mislocalization to the vacuole and impaired n-glycan processing in vivo. Our results suggest that sequence-specific features of the GnTI TMD are required for its interaction with a Golgi-resident adaptor protein or a specific lipid environment that likely promotes coat protein complexI-mediated retrograde transport, thus maintaining the steady-state distribution of GnTI in the cis/medial-Golgi of plants.
The Golgi apparatus has a central role in protein processing and transport along the secretory pathway. In the Golgi apparatus, secreted glycoproteins from animals and plants acquire complex glycan structures that furnish them with distinct functions (Helenius and Aebi, 2001; Strasser, 2016). The formation of complex n-glycans in the Golgi is carried out by specific glycosyltransferases and glycosidases that are asymmetrically distributed throughout the different Golgi cisternae (Dunphy and Rothman, 1983; Rabouille et al., 1995). The distinct, yet overlapping, distribution is a prerequisite for controlled glycosylation by spatial separation of glycan processing enzymes that compete for the same acceptor substrates. In mammals, diseases like congenital disorders of glycosylation are caused by missense mutations that affect trafficking and organization of Golgi-resident glycosylation enzymes (Fisher and Ungar, 2016). Despite their important role in maintenance of Golgi organization and function, the mechanisms for steady-state distribution of Golgi-resident glycosyltransferases and glycosidases are still largely unknown (Tu and Banfield, 2010). A better understanding of the Golgi organization is further complicated by the fact that the mode of cargo transport through the Golgi is still controversial in most organisms and different cell types (Glick and Luini, 2011; Ito et al., 2014). Sequence motifs that govern the intra-Golgi distribution can either function as a retention signal for a distinct Golgi subcompartment, a retrieval signal for retrograde transport, or a combination of both.
A common feature of Golgi-resident n-glycan processing enzymes is their type-II membrane protein topology with a short cytoplasmic tail, a single transmembrane domain (TMD), and a stem or linker region that orients the large catalytic domain into the Golgi lumen. Proposed models for Golgi retention and retrieval include oligomerization of glycosyltransferases and glycosidases with subsequent exclusion of large protein complexes from intra-Golgi transport (Machamer, 1991; Nilsson et al., 1993), TMD length-dependent sorting (Munro, 1995), lipid-based partitioning (Patterson et al., 2008), and cytoplasmic tail-dependent processes (Uliana et al., 2006; Petrosyan et al., 2015). An adaptor-mediated sorting mechanism involving binding to a specific amino acid motif within the cytoplasmic tail of glycosyltransferases and interaction with the coat protein complexI (COPI) retrograde transport system has been described in yeast (Saccharomyces cerevisiae) and mammals (Schmitz et al., 2008; Tu et al., 2008; Eckert et al., 2014). Yeast vacuolar protein sorting-associated protein74 (VPS74) acts as an adaptor protein that binds to the sequence motif in the cytoplasmic tail of late-Golgi glycosyltransferases and to COPI subunits to mediate the retrograde transport in the Golgi. Apart from VPS74 and its mammalian homologGolgi phosphoprotein3 (GOLPH3), which may have a similar function, no adaptor proteins for binding to cytoplasmic tails have been identified so far and for many Golgi-resident enzymes, like those involved in complex n-glycan processing in mammals or plants, the signals and mechanisms for their steady-state distribution are not well known. Noteworthy, an adapter-mediated mechanism has not been described in plants and VPS74/GOLPH3-like proteins appear absent. Interestingly, a recent study showed a direct binding of COPI subunits to an amino acid motif present in the cytoplasmic tail of several mammalian glycosyltransferases (Liu et al., 2018) indicating an adaptor-protein independent sorting. Golgi localization of plant n-glycan processing enzymes is typically mediated by sequence features within the n-terminal region comprising the cytoplasmic tail, the TMD, and the stem (CTS region) without any contribution from the catalytic domain (Saint-Jore-Dupas et al., 2006; Schoberer et al., 2010, 2013). However, within this N-terminal targeting region, a conserved sequence motif contributing to Golgi compartment-specific localization has yet to be revealed.
n-glycosylation is an essential posttranslational modification that is initiated in the endoplasmic reticulum (ER) by transfer of a preassembled oligosaccharide to Asn residues within the consensus sequence Asn-X-Ser/Thr (“X” can be any amino acid except Pro) on nascent proteins (Strasser, 2016). Upon transfer, the n-glycans are processed by specific α-glucosidases and α-mannosidases, and glycoproteins with oligomannosidic n-glycans exit the ER toward the Golgi complex. In the Golgi, additional mannose (Man) residues are removed by Golgi α-mannosidase I (Liebminger et al., 2009) and complex n-glycan formation is initiated by β1,2-n-acetylglucosaminyltransferaseI (GnTI, called “MGAT1” in mammals), which transfers a single GlcNAc residue to Man5GlcNAc2 (Man5). This is a critical step in the pathway because all further n-glycan processing reactions in the Golgi depend on the presence of the single GlcNAc added by GnTI (von Schaewen et al., 1993; Strasser et al., 1999). Importantly, the central function of GnTI in complex n-glycan formation is conserved in higher eukaryotes and GnTI deficiency is embryo-lethal in mammals (Ioffe and Stanley, 1994). GnTI-deficient Lotus japonicus displays severe growth defects and rice (Oryza sativa) lacking GnTI shows developmental phenotypes leading to early lethality (Fanata et al., 2013; Pedersen et al., 2017). Arabidopsis (Arabidopsis thaliana) lacking GnTI is viable, but less tolerant to abiotic stress and display severe growth defects when combined with mutations that affect plant cell wall formation (von Schaewen et al., 1993; Kang et al., 2008). In summary, these data highlight the importance of complex n-glycan initiation by GnTI for different biological processes across kingdoms.
In accordance with its n-glycan processing function, GnTI is mainly located in the cis/medial-Golgi in plants (Reichardt et al., 2007; Schoberer et al., 2010). In a previous study, we generated chimeric GnTI variants and analyzed their Golgi localization and functionality to identify regions for Golgi targeting and retrieval/retention (Schoberer et al., 2014). The results for tobacco (Nicotiana tabacum) GnTI demonstrated that the cytoplasmic tail, the stem region, and the catalytic domain are not involved in the intra-Golgi distribution of GnTI. By contrast, the TMD plays a pivotal role for its cis/medial-Golgi distribution. Here, we investigated the contribution of individual sequence motifs and amino acid residues in the TMD to Golgi localization and GnTI function. Tobacco and Arabidopsis GnTI (herein NtGnTI and AtGnTI, respectively) were analyzed to identify a common mechanism for their Golgi distribution. We identified a single amino acid residue conserved in the TMD of plant GnTI enzymes that plays an essential role in GnTI cis/medial-Golgi localization and n-glycan processing. Mutagenesis of this conserved residue altered the subcellular localization of GnTI and impaired its function when expressed under native-like conditions. In summary, our study reveals a motif that governs Golgi-resident enzyme distribution and provides insights into the subcellular localization mechanisms of Golgi-located type-II membrane proteins.
RESULTS
The Exoplasmic Half of the GnTI TMD Is Important for the Cis/Medial-Golgi Localization
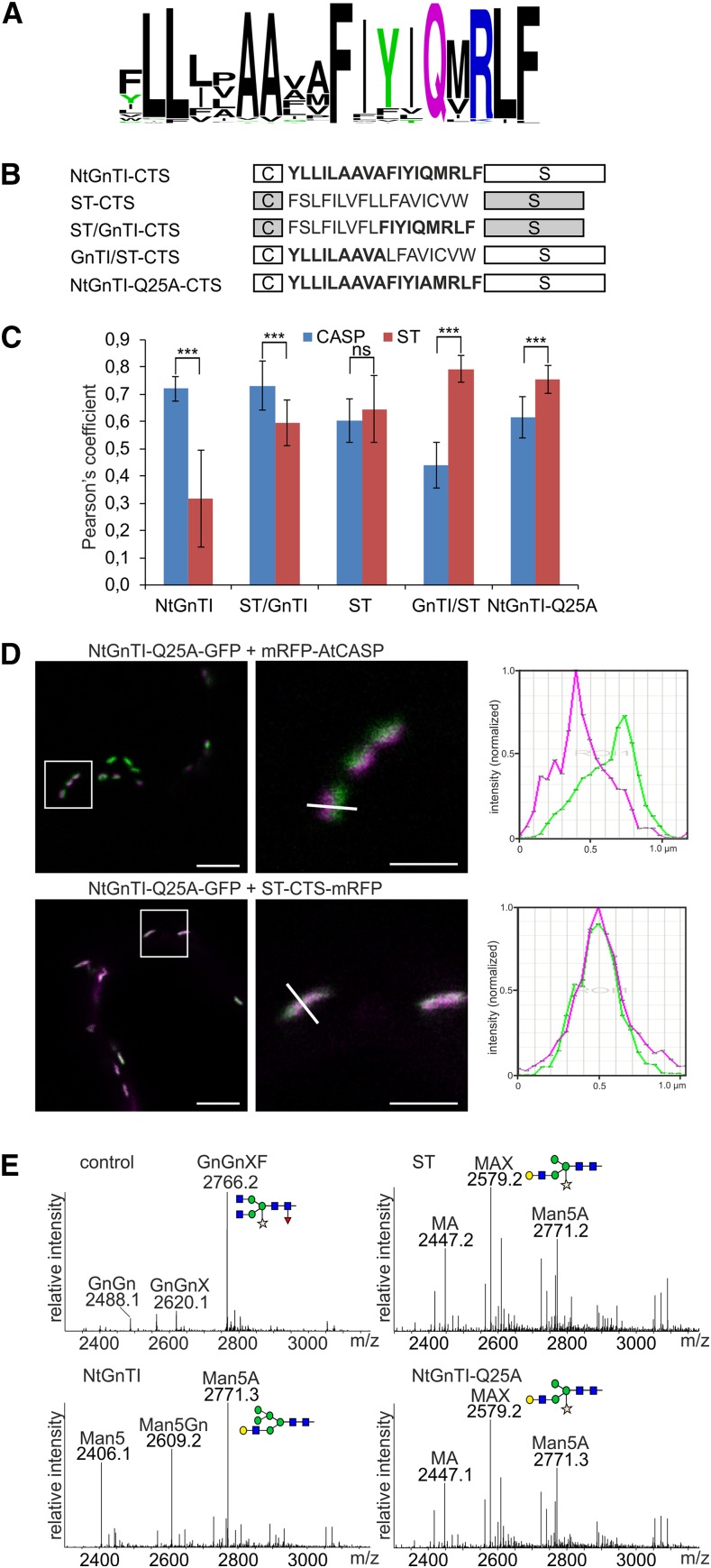
We compared the GnTI TMD sequences from different plant species to identify amino acid motifs that are involved in the cis/medial-Golgi distribution. The sequence alignment revealed the presence of several highly conserved amino acid residues, especially in the exoplasmic half of the TMD (Fig. 1A; Supplemental Fig. S1). Consequently, we exchanged the exoplasmic half of the TMD from the well-characterized trans Golgi marker rat α2,6-sialyltransferase (ST; Boevink et al., 1998) with nine exoplasmic amino acids from the 18-residue TMD of NtGnTI. The resulting chimeric CTS regions, ST/GnTI and GnTI/ST (Fig. 1B; Supplemental Fig. S2), were C-terminally fused to the NtGnTI catalytic domain and green fluorescent protein (GFP), transiently expressed in Nicotiana benthamiana leaves and examined using a confocal microscope. Colocalization was carried out with the β1,2-galactosyltransferase (MUR3), a xyloglucan synthesizing enzyme that has been associated with medial-Golgi cisternae (Chevalier et al., 2010). As expected, both constructs harboring the chimeric CTS regions displayed Golgi localization (Supplemental Fig. S3). To assess the degree of colocalization in the Golgi, the Pearson’s correlation coefficient was determined for colocalization with the cis-Golgi protein Arabidopsis CCAAT-displacement protein alternatively spliced product (AtCASP; Osterrieder et al., 2010; Schoberer et al., 2010) fused to monomeric red fluorescent protein (mRFP) and the trans-Golgi marker ST-CTS-mRFP (Fig. 1C; Supplemental Table S1). The experiment was carried out with the CTS regions lacking the catalytic domains to enhance their expression. The colocalization analysis revealed a similar correlation for ST/GnTI-CTS-GFP and NtGnTI-CTS-GFP as well as for GnTI/ST-CTS-GFP and ST-CTS-GFP, indicating that the nine exoplasmic amino acids from the GnTI TMD contain a sequence motif for the concentration in a distinct Golgi subcompartment.
Figure 1.
The exoplasmic half of the NtGnTI TMD contains a signal for cis/medial-Golgi localization. A, Sequence logo illustration of the degree of amino acid conservation within the TMD of plant GnTI sequences (74 sequences in total). B, Schematic illustration of the NtGnTI, ST, ST/GnTI, and GnTI/ST CTS regions. The amino acid sequence of the TMD is shown for each CTS region. “C”: cytoplasmic tail; “S”: stem region. Sequences corresponding to the NtGnTI TMD are highlighted in bold. C, Colocalization analysis. NtGnTI-CTS-GFP, ST/GnTI-CTS-GFP, ST-CTS-GFP, and GnTI/ST-GFP were coexpressed with mRFP-AtCASP (CASP) or ST-CTS-mRFP (ST; data are the mean ± sd, n > 10 confocal images). Asterisks indicate a significant difference between CASP and ST localization (P ≤ 0.001, two-tailed t test: two-sample assuming equal variances); ns, not significant (P > 0.05). D, Colocalization of NtGnTI-Q25A-GFP with mRFP-AtCASP or with ST-CTS-mRFP. Left images, scale bars = 5 μm; right images, scale bars = 1 μm. Magnified images of the boxed region from the left images are shown in the right images. Graphs show relative fluorescence intensities (NtGnTI-Q25A-GFP in green and mRFP-AtCASP or ST-CTS-mRFP in magenta) along the white lines (from the right images). E, Glycan processing assay. NtGnTI-Q25A-GALT or the indicated controls were coexpressed with a glycoprotein reporter. LC-ESI-MS analysis of the glycopeptide “EEQYNSTYR” derived from the glycoprotein is shown. Reporter without GALT (control), reporter coexpressed with ST-GALT (ST), NtGnTI-GALT (NtGnTI), or NtGnTI-Q25A-GALT (NtGnTI-Q25A). The schematic presentation of the major n-glycan structure is given. The symbols are drawn according to the nomenclature from the Consortium for Functional Glycomics. For further details on the abbreviation of the n-glycan structures, see Supplemental Figure S4.
To obtain further support for our finding, we fused the ST/GnTI-CTS and GnTI/ST-CTS regions to the catalytic domain of human β1,4-galactosyltransferase (GALT) and performed an n-glycan processing assay. In this assay, the coexpression of GALT with a secreted glycoprotein reporter results in the transfer of β1,4-Gal to the nonreducing end of n-glycans on the reporter (Schoberer et al., 2014). Dependent on the Golgi compartmental localization of the chimeric GALT, a different n-glycan pattern is obtained due to competition of GALT with endogenous n-glycan processing enzymes (Supplemental Fig. S4). Thus, the n-glycan processing assay allows cis/medial-Golgi-localized GALT (leading to incompletely processed n-glycans) to be distinguished from medial/trans-Golgi-localized GALT (higher amount of completely processed n-glycans). Mass spectrometry (MS)-based analysis of a glycopeptide derived from the reporter revealed incompletely processed n-glycans for ST/GnTI-CTS and a substantial amount of completely processed n-glycans for GnTI/ST-CTS (Supplemental Fig. S5), providing additional evidence for a role of the exoplasmic TMD half in intra-Golgi localization.
We have previously shown that a trans-Golgi targeted GnTI carrying the ST-CTS region fails to complement the complex n-glycan processing defect of GnTI-deficient Arabidopsis plants (Schoberer et al., 2014). Here, we fused the chimeric CTS regions with the exchanged exoplasmic TMD domains to the AtGnTI catalytic domain. GFP-fused AtGnTI variants were expressed in Arabidopsis gntI T-DNA insertional mutants under the control of a weak endogenous AtGnTI promoter and the processing to complex n-glycans was analyzed by immunoblotting. Consistent with an effect on cis/medial-Golgi localization, GnTI/ST-AtGnTI-GFP carrying the exoplasmic TMD region from ST did not restore complex n-glycan formation (Supplemental Fig. S6). However, eight out of 12 tested transgenic ST/GnTI-AtGnTI-GFP lines displayed considerable amounts of complex n-glycans, indicating relocation of the protein to an earlier Golgi compartment where it initiated complex n-glycan processing. Taken together, our results show that an amino acid motif in the exoplasmic half of the NtGnTI TMD contains a signal for efficient steady-state cis/medial-Golgi localization.
A Single Amino Acid Residue in the NtGnTI TMD Is Crucial for Cis/Medial-Golgi Localization
To further characterize the sequence motif in the exoplasmic half of the NtGnTI TMD, we generated another chimeric GALT variant where the LFAV tetrapeptide sequence from ST was replaced by the FIYIQ amino acid motif from NtGnTI (STFIYIQ-GALT) and carried out the n-glycan processing assay. Coexpression of STFIYIQ-GALT resulted in the generation of incompletely processed n-glycans (Supplemental Fig. S5), indicating that the FIYIQ-motif plays a role in NtGnTI cis/medial-Golgi localization. One of the two fully conserved residues in this motif is the Gln at position 25 (Fig. 1A). We replaced this Gln (Q25) with Ala and analyzed the localization of NtGnTI-Q25A fused to GFP or mRFP in N. benthamiana leaf epidermal cells. NtGnTI-Q25A was found in Golgi bodies as well as other intracellular compartments indicating that the subcellular localization was changed (Supplemental Fig. S3). Colocalization of NtGnTI-Q25A-GFP with mRFP-AtCASP and ST-CTS-mRFP indicated a shift in the Golgi localization toward the trans-Golgi (Fig. 1D). The N-glycan processing assay revealed the presence of processed n-glycans when NtGnTI-Q25A-GALT was coexpressed with the glycoprotein reporter and the n-glycan profile was comparable to that derived from ST-GALT (Fig. 1E).
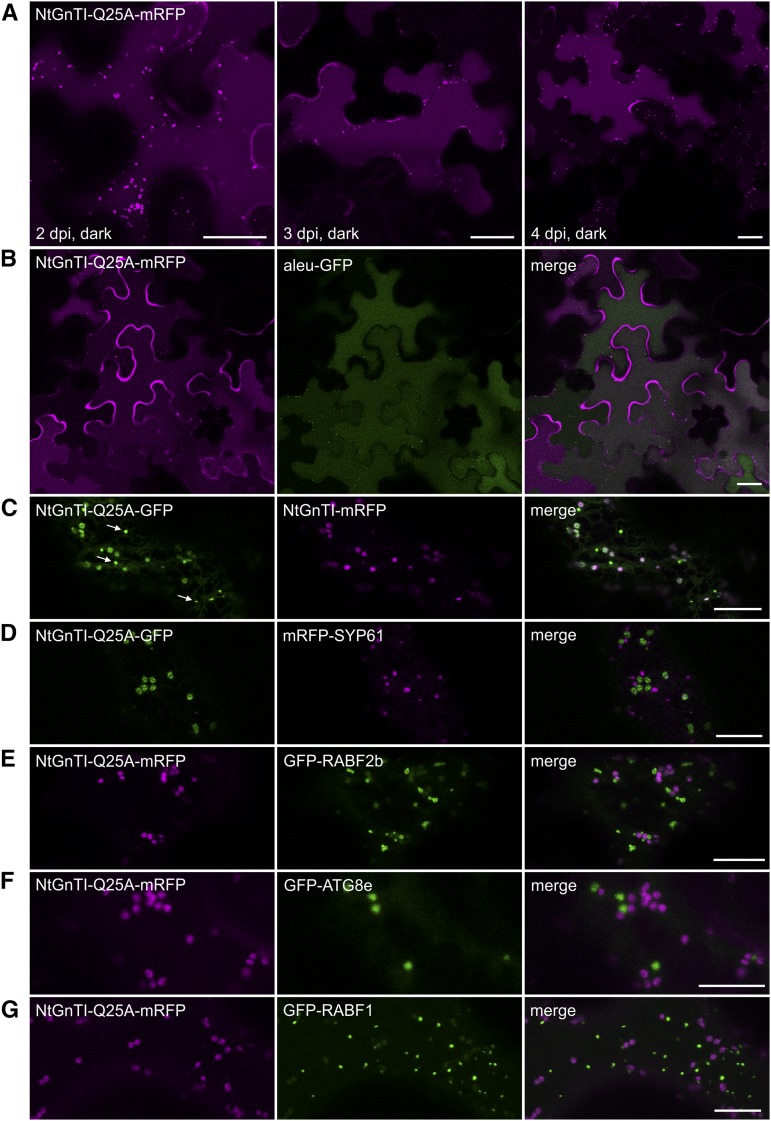
In addition to the accumulation in the trans-Golgi, GFP- or mRFP-tagged NtGnTI-Q25A was observed in the vacuole (Fig. 2A; Supplemental Fig. S3), which was clearly visible 3–5 d after infiltration. Colocalization with aleu-GFP (Humair et al., 2001) confirmed targeting to the lytic vacuole (Fig. 2B). No ER localization was observed for NtGnTI-Q25A fusion proteins, but accumulation of fluorescence in the apoplast was occasionally observed, indicating the secretion of full-length NtGnTI-Q25A or a degradation product. In addition to vacuolar and trans-Golgi localization, we observed smaller puncta in NtGnTI-Q25A-expressing cells that did not represent typical Golgi bodies (Fig. 2C). Additional colocalization experiments with different subcellular markers were carried out to identify these small puncta. However, the cellular structures overlapped with neither the TGN marker mRFP-SYP61 (Fig. 2D; Sanderfoot et al., 2001), the early endosome marker GFP-RABF2b (ARA7; Fig. 2E; Kotzer et al., 2004), the autophagosome marker GFP-ATG8e (Fig. 2F; Contento et al., 2005), nor the late endosome/multivesicular body marker GFP-RABF1 (ARA6; Fig. 2G; Ueda et al., 2001). These data show that the Gln residue confers efficient cis/medial-Golgi localization of NtGnTI. The mutated NtGnTI-Q25A, on the other hand, accumulates in the trans-Golgi and substantial amounts are released to post-Golgi compartments, including the vacuole by an unknown trafficking route.
Figure 2.
NtGnTI-Q25A locates in the Golgi, small puncta, and the vacuole. Confocal microscopy analysis was performed on N. benthamiana leaf epidermal cells expressing NtGnTI-Q25A fused to mRFP or GFP and different subcellular markers. A, Localization of NtGnTI-Q25A-mRFP was monitored at different time points postinfiltration (2, 3, and 4 dpi). B, Colocalization with aleu-GFP (vacuolar marker). C, Colocalization of NtGnTI-Q25A-GFP with NtGnTI-mRFP reveals the labeling of small punctate structures (white arrows) that are distinct from NtGnTI-labeled Golgi stacks. D, Colocalization with mRFP-SYP61 (TGN-marker). E, Colocalization with GFP-RABF2b (early endosome marker). F, Colocalization with GFP-ATG8e (autophagosome marker). G, Colocalization with GFP-RABF1 (late endosome/multivesicular body marker). Scale bars = 25 µm (A and B) and 10 µm (C–G).
Arabidopsis GnTI-Q23A Fails to Complement GnTI-Deficient Plants
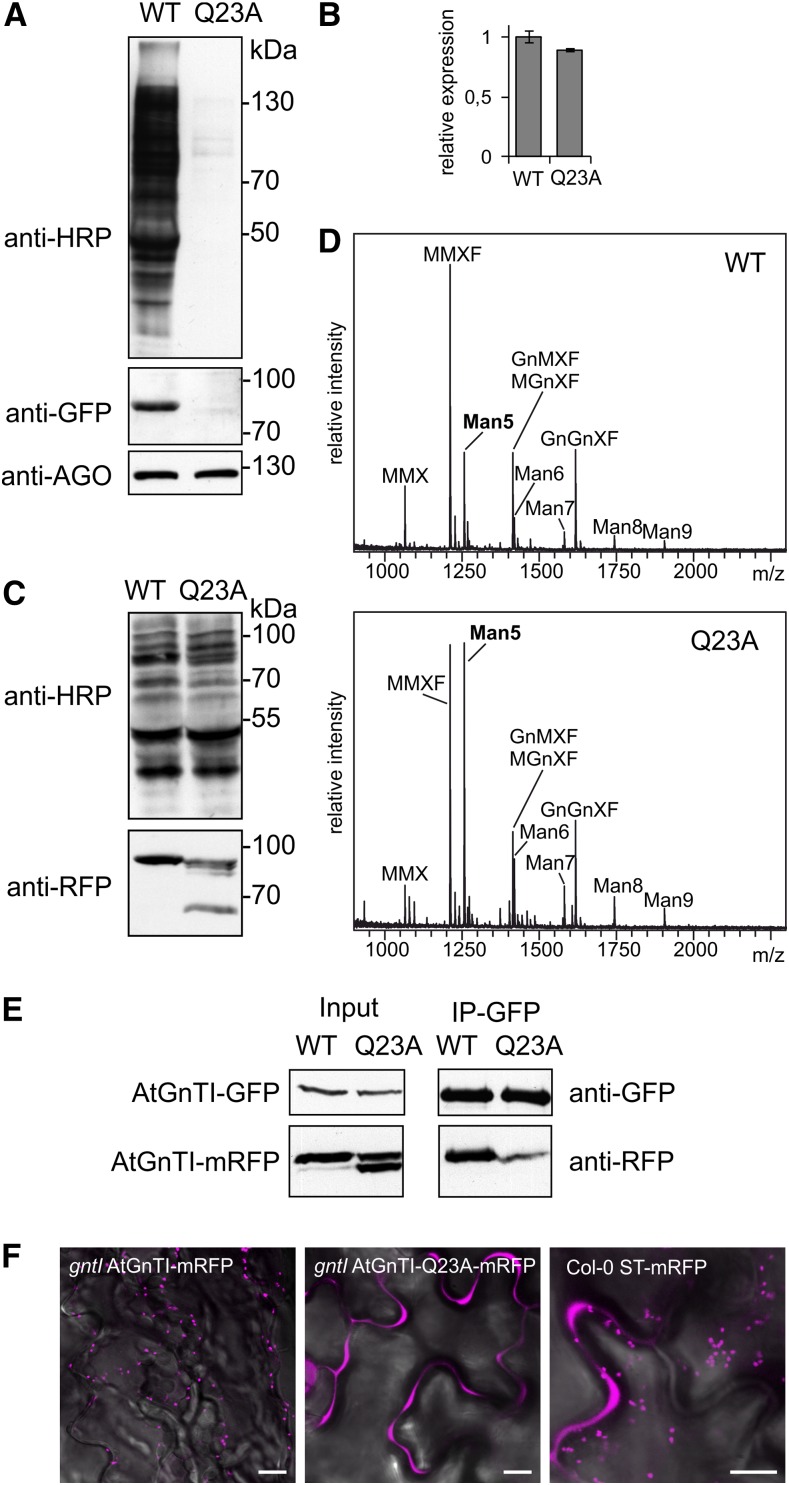
To investigate the functional relevance of the identified Gln residue within the TMD of GnTI, we expressed GFP- and mRFP-tagged AtGnTI and the AtGnTI-Q23A variant with the conserved Gln (at position 23 in AtGnTI) changed to an Ala in the gntI mutant. A functional AtGnTI in the cis/medial-Golgi will restore the n-glycan processing that can be analyzed by immunoblotting using antibodies directed against Golgi-processed n-glycans carrying β1,2-Xyl and core α1,3-Fuc (Strasser et al., 2004). When expressed under the control of the weak AtGnTI promoter, complex n-glycans were detected in gntI plants expressing AtGnTI-GFP, but not in AtGnTI-Q23A-GFP–expressing plants (Fig. 3A). AtGnTI-GFP could be found on immunoblots, whereas AtGnTI-Q23A-GFP was hardly detectable. Comparison of the transcript levels between the two AtGnTI variants by reverse transcription quantitative PCR (RT-qPCR) did not reveal considerable variation in mRNA levels (Fig. 3B), suggesting that the difference in protein abundance is caused by a decreased stability of the AtGnTI-Q23A-GFP protein. The same result was obtained for complementation of Arabidopsis cgl1 plants that carry a missense mutation in the AtGnTI gene (von Schaewen et al., 1993; Strasser et al., 2005) or when mRFP-tagged AtGnTI variants were used for complementation of gntI (Supplemental Fig. S7). Matrix-assisted laser-desorption ionization-time of flight (MALDI-TOF) MS analysis of n-glycans from gntI expressing AtGnTI-GFP under the control of the weak endogenous AtGnTI promoter showed partial formation of complex n-glycans. By contrast, no peaks corresponding to complex n-glycans were detectable for AtGnTI-Q23A-GFP (Supplemental Fig. S8). Expression of AtGnTI-mRFP or AtGnTI-Q23A-mRFP under the control of the constitutive ubiquitin10 (UBQ10) promoter restored the complex n-glycan formation in gntI plants (Fig. 3C). However, in gntI expressing the mutant AtGnTI-Q23A variant, MALDI-TOF MS analysis revealed the presence of considerable amounts of unprocessed Man5, which is the major n-glycan in AtGnTI-deficient plants (Fig. 3D; Supplemental Fig. S8; von Schaewen et al., 1993; Strasser et al., 2005). Thus, even the overexpression of the AtGnTI-Q23A variant could not fully complement the n-glycan processing defect of gntI, which would result in reduced levels of Man5 and more of the truncated n-glycan carrying β1,2-Xyl and core α1,3-Fuc. This finding is fully consistent with the observed mistargeting due to the disturbance of the conserved sequence motif within the GnTI TMD.
Figure 3.
AtGnTI-Q23A fusion protein expression does not restore the n-glycan processing defect of gntI. A, Immunoblot analysis of gntI AtGnTI:AtGnTI-GFP (WT) and gntI AtGnTI:AtGnTI-Q23A-GFP (Q23A) seedlings with anti-HRP to detect plant complex n-glycans, with GFP antibodies (anti-GFP) to detect the AtGnTI-GFP fusion protein and with antibodies against ARGONAUTE 1 (anti-AGO) used as a loading control. B, RT-pPCR analysis of transcripts from gntI AtGnTI:AtGnTI-GFP (WT) and gntI AtGnTI:AtGnTI-Q23A-GFP (Q23A) seedlings. PCR for the transgene expression was carried out with GFP-specific primers and normalized to UBQ5 transcript expression. Data represent mean values ± se, n = 3. C, Immunoblot analysis of protein extracts from gntI UBQ10:AtGnTI-mRFP (WT) or gntI UBQ10:AtGnTI-Q23A-mRFP (Q23A) with anti-HRP and anti-RFP. D, n-glycan analysis of complemented gntI expressing UBQ10:AtGnTI-mRFP (WT) or UBQ10:AtGnTI-Q23A-mRFP (Q23A) by MALDI-MS. E, Co-immunoprecipitationof AtGnTI (WT) and AtGnTI-Q23A (Q23A). UBQ10:AtGnTI-GFP coexpressed with UBQ10:AtGnTI-mRFP (WT) or UBQ10:AtGnTI-Q23A-mRFP (Q23A) was purified with GFP-trap beads from N. benthamiana and the copurification of mRFP-fusion proteins was monitored by immunoblotting with mRFP antibodies. WT, wild type. F, Localization of AtGnTI-mRFP and AtGnTI-Q23A-mRFP expressed in Arabidopsis seedlings under the control of the UBQ10 promoter and ST-mRFP expressed under the control of the CaMV35S promoter. Scale bars = 10 µm.
Immunoblot analysis of AtGnTI-mRFP stably expressed under the control of the UBQ10 promoter in gntI revealed a single band of the expected size of ∼77 kD for the fusion protein. By contrast, the AtGnTI-Q23A variant displayed 3–4 bands (Fig. 3C). The upper one corresponds to the size of the full-length AtGnTI-mRFP protein, whereas the other 2–3 bands represent faster migrating variants of different sizes suggesting the presence of degradation products. In a previous study, we observed that GnTI forms a homodimer that requires a sequence domain from the stem region (Schoberer et al., 2013, 2014). Here, we tested whether mistargeted AtGnTI-Q23A can still interact with the nonmutated AtGnTI. Both proteins were coexpressed in N. benthamiana leaf epidermal cells and subjected to coimmunoprecipitation analysis. In contrast to wild-type AtGnTI, a substantially decreased amount of AtGnTI-Q23A was copurified (Fig. 3E). Notably, only the band representing full-length AtGnTI interacted, whereas the faster migrating bands from AtGnTI-Q23A were not copurified—providing potential evidence that they represent spatially separated variants from post-Golgi compartments. Förster resonance energy transfer-fluorescence lifetime imaging (FRET-FLIM) analysis of transiently expressed NtGnTI and NtGnTI-Q25A fusion proteins showed that the FRET efficiency determined by FLIM was also reduced for the wild-type/mutant NtGnTI combination compared to the homodimer formation of wild-type NtGnTI (Table 1). Confocal microscopy of transgenic seedlings confirmed the Golgi localization of AtGnTI-mRFP. AtGnTI-Q23A-mRFP–expressing seedlings hardly displayed any Golgi localization, but showed a fluorescence signal in the apoplast that may be derived from the cleavage and secretion of mRFP. A similar secretion has been described for the trans-Golgi marker ST-mRFP (Runions et al., 2006), but compared to AtGnTI-Q23A-mRFP, the effect is less pronounced (Fig. 3F).
Table 1. FRET efficiency determined by FLIM.
A minimum decrease of the average excited-state fluorescence lifetime of the donor molecule by 0.20 ns or 8% in the presence of the acceptor molecule was considered relevant to indicate interaction. τD, lifetime of the donor in the absence of the acceptor; τDA, lifetime of the donor in the presence of the acceptor; Δτ, lifetime contrast (τD-τDA); E, FRET efficiency calculated according to (1 − [τD − τDA]) × 100; ns, nanosecond; n, number of Golgi bodies analyzed.
| Donor | Acceptor | τD ± sd (ns) | τDA ± sd (ns) | Δτ (ns) | E (%) |
|---|---|---|---|---|---|
| NtGnTI-CTS-GFP | NtGnTI-CTS-mRFP | 2.44 ± 0.06 (n = 412) | 2.08 ± 0.09 (n = 204) | 0.36 | 14.56 |
| NtGnTI-CTS-GFP | ST-CTS-mRFP | 2.44 ± 0.06 (n = 412) | 2.36 ± 0.06 (n = 238) | 0.08 | 3.35 |
| NtGnTI-CTS-GFP | NtGnTI-CTS-Q25A-mRFP | 2.44 ± 0.06 (n = 412) | 2.37 ± 0.06 (n = 273) | 0.07 | 2.89 |
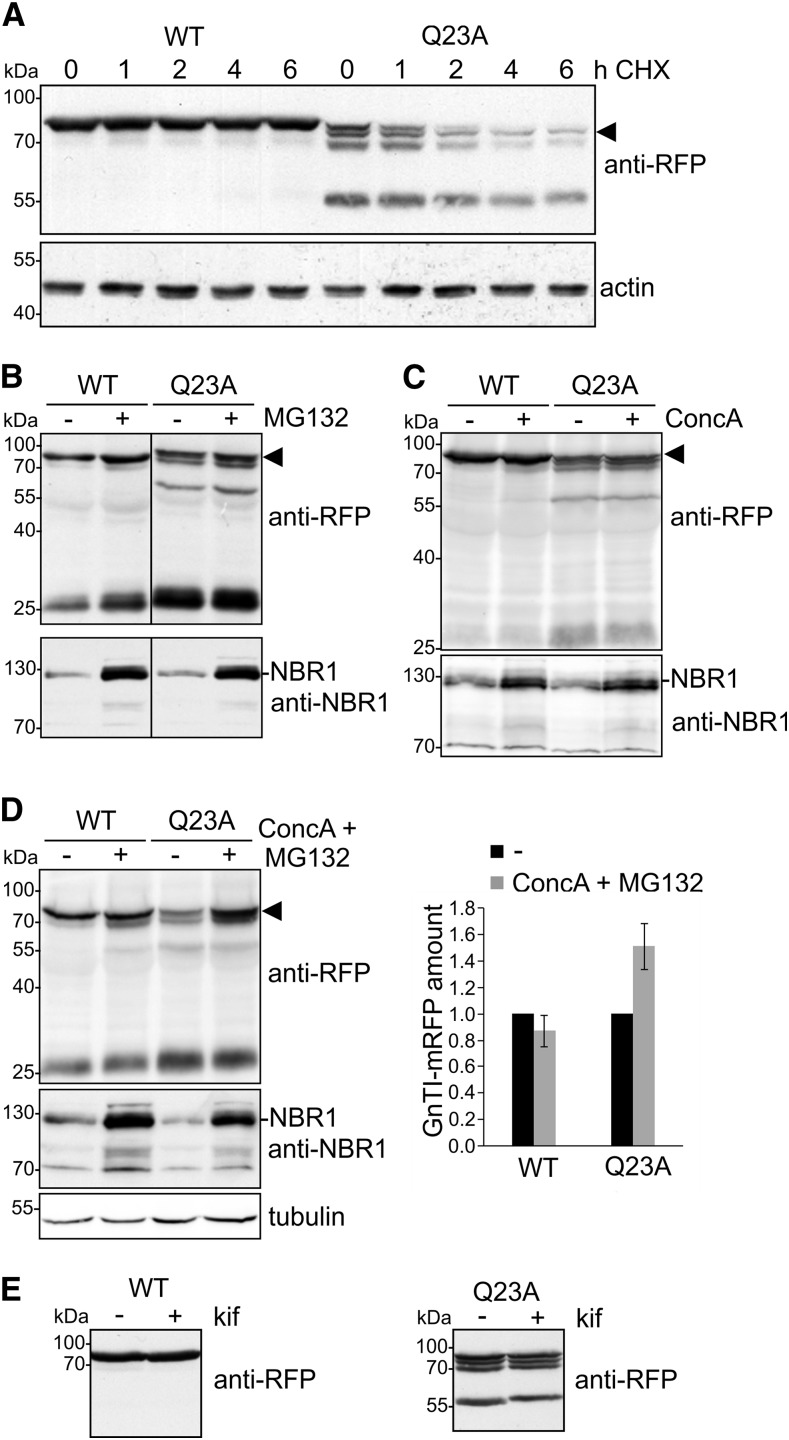
To investigate the fate of the mutant variant in more detail, 10-d–old seedlings were treated with cycloheximide (CHX) and the AtGnTI-mRFP protein level was analyzed at different time points by immunoblotting. AtGnTI-mRFP levels were stable during the chase period. The AtGnTI-Q23A-mRFP levels, on the other hand, decreased over time, indicating a faster clearance (Fig. 4A). Moreover, we treated Arabidopsis seedlings with the proteasomal inhibitor MG132 (Fig. 4B) as well as with the vacuolar H+-ATPase inhibitor concanamycin A (Dettmer et al., 2006) to see whether the discrete degradation products were generated in an acidic compartment like the vacuole (Fig. 4C). By increasing the pH in the organelle, the catalytic activity of proteases is diminished, which may result in the accumulation of proteins targeted for vacuolar degradation (Chamberlain et al., 2008). Pharmacological inhibition with MG132 or concanamycin A did not result in substantial changes in the pattern of AtGnTI-Q23A-mRFP degradation products. However, incubation of Arabidopsis seedlings in a combination of MG132 and concanamycin A increased the amounts of the intact AtGnTI-Q23A-mRFP protein (Fig. 4D). By contrast, the ER-associated degradation inhibitor kifunensine did not stabilize AtGnTI-Q23A-RFP (Fig. 4E).
Figure 4.
AtGnTI-Q23A-mRFP degradation is blocked in the presence of both MG132 and concanamycin A. A, CHX-degradation assay. Nine-d–old Arabidopsis gntI UBQ10:AtGnTI-mRFP (WT) and gntI UBQ10:AtGnTI-Q23A-mRFP (Q23A) seedlings were incubated for the indicated time in 100 µg/mL of CHX and subjected to SDS-PAGE and immunoblotting. The intact fusion protein is marked by an arrowhead. Detection of actin was used as a control. B–D, gntI UBQ10:AtGnTI-mRFP (WT) and gntI UBQ10:AtGnTI-Q23A-mRFP (Q23A) seedlings were incubated for 17 h with 20 μM of MG132 (B), with 1 μM of concanamycin A (ConcA, C), or with 1 μM of concanamycin A and 20 μM of MG132 (D). AtGnTI expression was monitored with anti-RFP. NBR1 detection served as a control for the inhibitor treatment (increased NBR1 signal) and tubulin as a loading control. The quantitation shows the relative GnTI-mRFP amount normalized to tubulin expression. Values are means ± se (n = 4 independent experiments). E, gntI UBQ10:AtGnTI-mRFP (WT) and gntI UBQ10:AtGnTI-Q23A-mRFP (Q23A) seedlings were incubated for 24 h with 20 µM of the class I α-mannosidase inhibitor kifunensine (kif) and proteins were subjected to SDS-PAGE and immunoblotting with anti-RFP antibodies.
A Polar or Positively Charged Amino Acid Is Required for Proper Golgi Targeting and In Vivo n-Glycan Processing
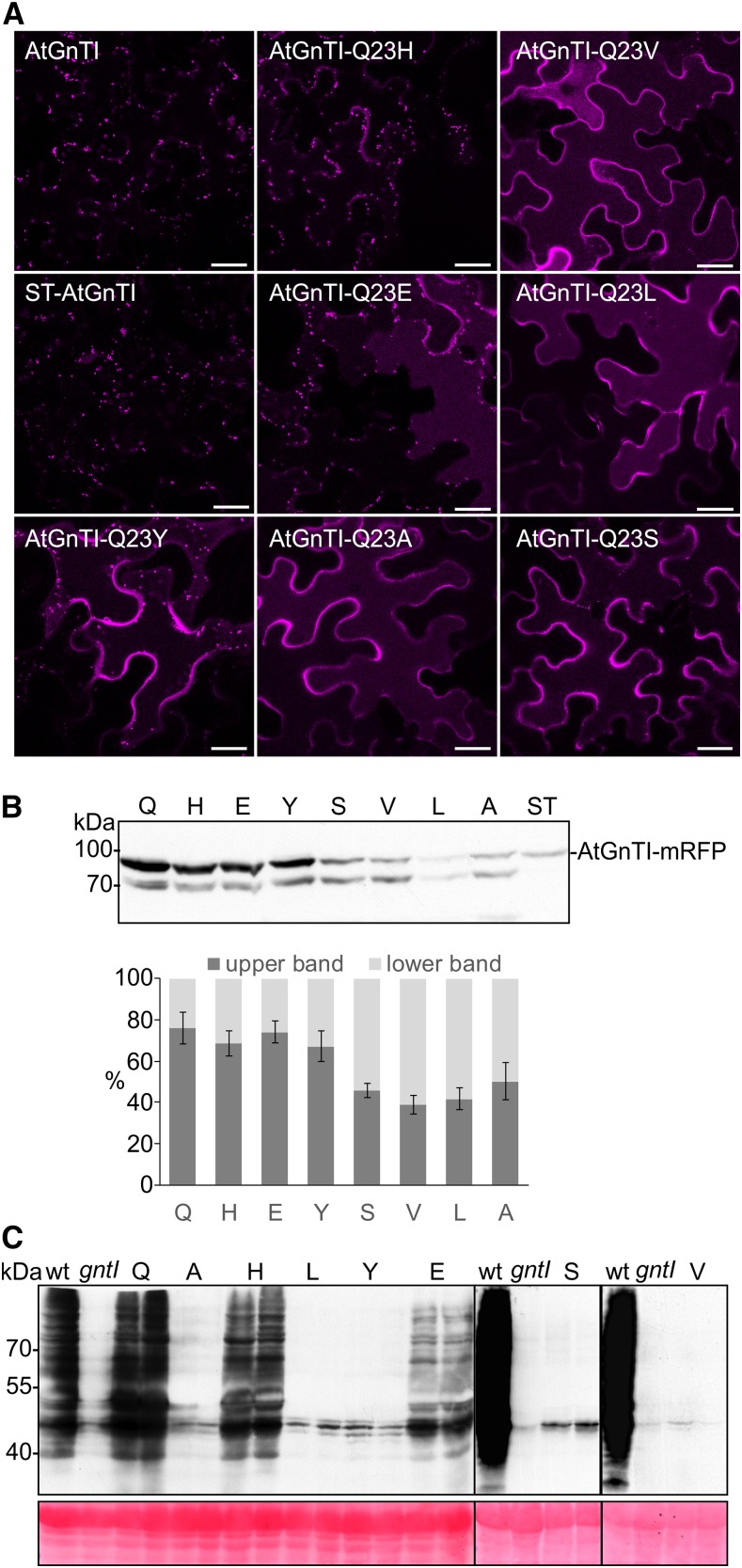
Different features of the TMD have been suggested to play a role for the subcellular localization of type-II membrane proteins including the TMD length, the presence of polar residues in the exoplasmic region of the leaflet, or changes in the average amino acid volume in the exoplasmic half (Munro, 1995; Sharpe et al., 2010; Quiroga et al., 2013). To define the signal for proper AtGnTI localization in more detail, we analyzed the contribution of different amino acid residues at this position to GnTI localization and processing of complex n-glycans. We performed site-directed mutagenesis and generated mutants where Q23 from AtGnTI was replaced by His, Leu, Glu, Tyr, Val, and Ser. Confocal microscopy of mRFP-fusion proteins expressed under the control of the UBQ10 promoter showed that AtGnTI-Q23V, AtGnTI-Q23L, and AtGnTI-Q23S behaved like AtGnTI-Q23A and displayed mistargeting to post-Golgi compartments. AtGnTI-Q23H showed mainly Golgi localization similar to that of AtGnTI. AtGnTI-Q23E and AtGnTI-Q23Y displayed Golgi localization as well as post-Golgi distribution (Fig. 5A; Supplemental Fig. S9). In agreement with altered localization, an increase of the faster migrating degradation product was observed for various AtGnTI mutants (Fig. 5B). Together, these data suggest that a polar amino acid residue with a long side chain (Gln) or a positively charged residue like His is tolerated at this particular position within the GnTI TMD, whereas hydrophobic residues with small (e.g. Ala) or large volumes (e.g. Leu) are not tolerated (Supplemental Fig. S10). To determine the effect of the different residues on in vivo n-glycan processing, transgenic Arabidopsis gntI expressing the different mutant variants under the control of the AtGnTI promoter were generated. In agreement with our localization data, only AtGnTI-Q23H-RFP displayed a similar degree of complex n-glycan formation like AtGnTI-RFP. Other mutants did not complement the defect at all or resulted only in partial complementation (e.g. AtGnTI-Q23E; Fig. 5C). Likewise, substitution of adjacent highly conserved amino acids (F19A, M24I, and R25A) did not obviously affect the subGolgi distribution of AtGnTI and resulted in gntI complementation (Supplemental Fig. S11).
Figure 5.
AtGnTI variants with an altered amino acid at position 23 are mistargeted in plants and do not complement the gntI n-glycan processing defect. A, Confocal microscopy analysis in N. benthamiana leaf epidermal cells of transiently expressed AtGnTI-Q23-mRFP mutants. Confocal images were acquired 48 h after infiltration. B, Transiently expressed AtGnTI-Q23-mRFP mutants analyzed by immunoblotting using an anti-mRFP antibody. The quantitation shows the relative amount of the upper (intact AtGnTI-mRFP) and the lower (AtGnTI-mRFP degradation product) bands. Values are means ± sd (n = 4 independent experiments. The single amino acid labeling indicates the substituted amino acid at position 23. C, Immunoblot analysis of transgenic gntI plants expressing the different AtGnTI-Q23-mRFP variants under the control of the endogenous AtGnTI promoter. Protein extracts from leaves were analyzed with antibodies against complex plant n-glycans (anti-HRP). Two independent lines are shown for each AtGnTI-Q23-mRFP variant, Col-0 (wild type), and gntI protein extracts were included as controls. Ponceau S staining of the membrane was used as a loading control.
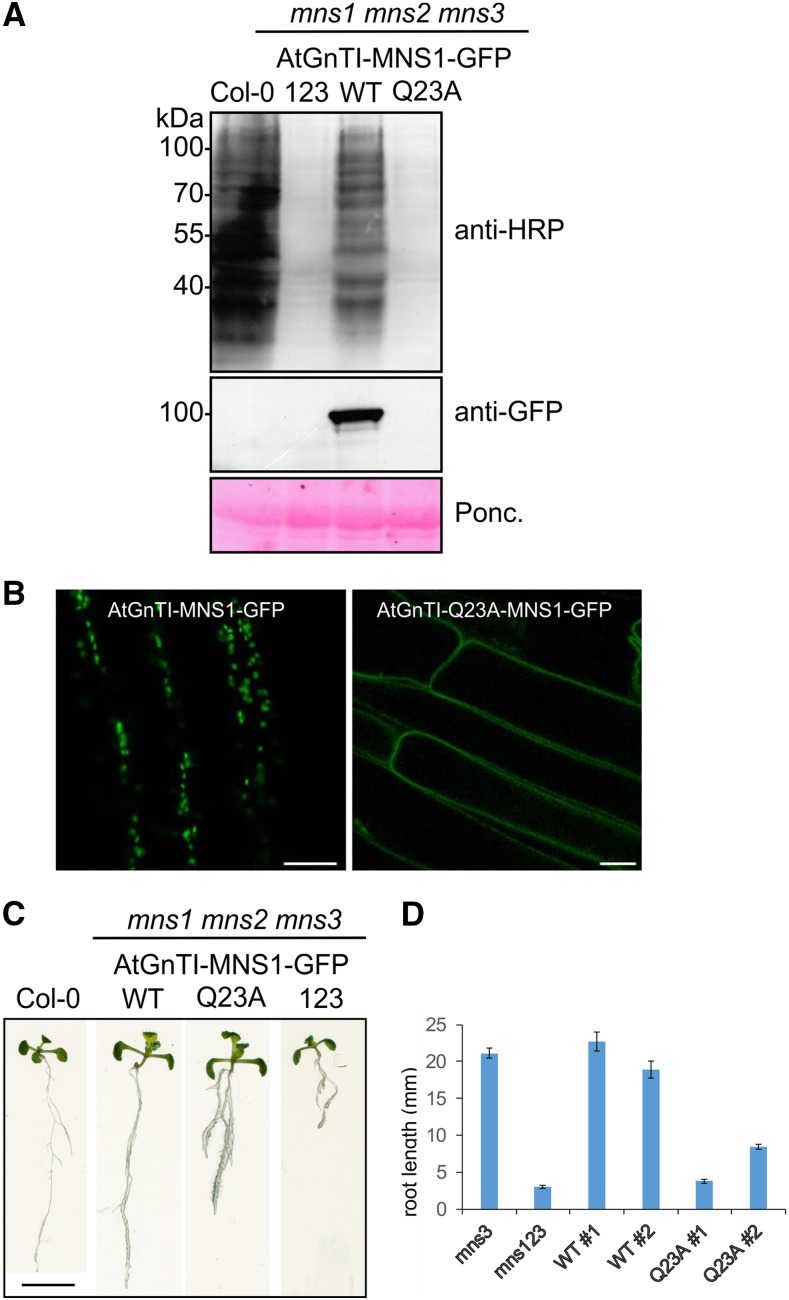
The Arabidopsis Golgi α-Mannosidase MNS1 Is Mistargeted and Nonfunctional when Fused to the AtGnTI-CTS-Q23A Domain
The conserved Gln in the TMD is important for the cis/medial-Golgi localization of GnTI. To see whether other n-glycan processing enzymes are mistargeted when their CTS region is exchanged with the mutated GnTI domain, we fused the AtGnTI CTS region to the catalytic domain of the Arabidopsis Golgi α-mannosidase1 (MNS1), which trims Man residues from n-glycans in the Golgi and acts upstream of GnTI by converting Man8GlcNAc2 to Man5 (Liebminger et al., 2009). We generated chimeric AtGnTI-CTS-MNS1-GFP and AtGnTI-CTS-Q23A-MNS1-GFP constructs, where the CTS region from GnTI is fused to the catalytic domain of MNS1 and GFP. The chimeric proteins were expressed under the control of the endogenous MNS1 promoter in the mns1 mns2 mns3 triple mutant that generates mainly Man9GlcNAc2 structures. Restoration of MNS1 activity in the cis/medial-Golgi of mns1 mns2 mns3 leads to the formation of complex n-glycans (Liebminger et al., 2009). In contrast to AtGnTI-CTS-MNS1-GFP, the MNS1 variant with the Q23A substitution in the AtGnTI TMD failed to restore complex n-glycan formation (Fig. 6A) and no Golgi localization was detected in root cells (Fig. 6B). We previously showed that the mns1 mns2 mns3 mutant displays a severe root-growth phenotype due to the block in n-glycan processing (Liebminger et al., 2009). Expression of MNS1:AtGnTI-CTS-MNS1-GFP in mns1 mns2 mns3 led to an almost wild-type–like root growth of seedlings whereas the MNS1 variant harboring the GnTI targeting region with the mutated Gln displayed aberrantly swollen and truncated roots (Fig. 6C). The difference was even more pronounced when seeds were grown on 4% (w/v) Suc, which enhances the phenotype (Fig. 6D; Supplemental Fig. S12). These results highlight the importance of proper Golgi localization mediated by the Gln-containing sequence motif in the GnTI TMD.
Figure 6.
Mistargeted MNS1-GFP does not complement the n-glycan processing defect and root growth phenotype of mns1 mns2 mns3. MNS1:AtGnTI-MNS1-GFP and MNS1:AtGnTI-Q23A-MNS1-GFP were expressed in the Arabidopsis mns1 mns2 mns3 mutant. A, Protein extracts were analyzed from wild-type (Col-0), mns1 mns2 mns3 (123), mns1 mns2 mns3 complemented with MNS1:AtGnTI-MNS1-GFP (WT) or MNS1:AtGnTI-Q23A-MNS1-GFP (Q23A) by SDS-PAGE and immunoblotting using antibodies against complex plant n-glycans (anti-HRP) and GFP. Ponceau S (Ponc.) staining of the membrane was used as a loading control. B, Confocal microscopy of Arabidopsis roots expressing the indicated constructs. Scale bars = 10 µm. C, Phenotypic comparison of 12-d–old Col-0, mns1 mns2 mns3 (123), mns1 mns2 mns3 complemented with MNS1:AtGnTI-MNS1-GFP (WT) or MNS1:AtGnTI-Q23A-MNS1-GFP (Q23A) seedlings grown on 0.5× Murashige and Skoog supplemented with 1% Suc. Scale bar = 1 cm. D, Quantification of the root length from 8-d–old seedlings grown on 0.5× Murashige and Skoog supplemented with 4% Suc. For WTand Q23A, the data from two independent lines are shown. The mns3 single and the mns1 mns2 mns3 riple mutant are included as controls. Data represent means ± se (n > 50).
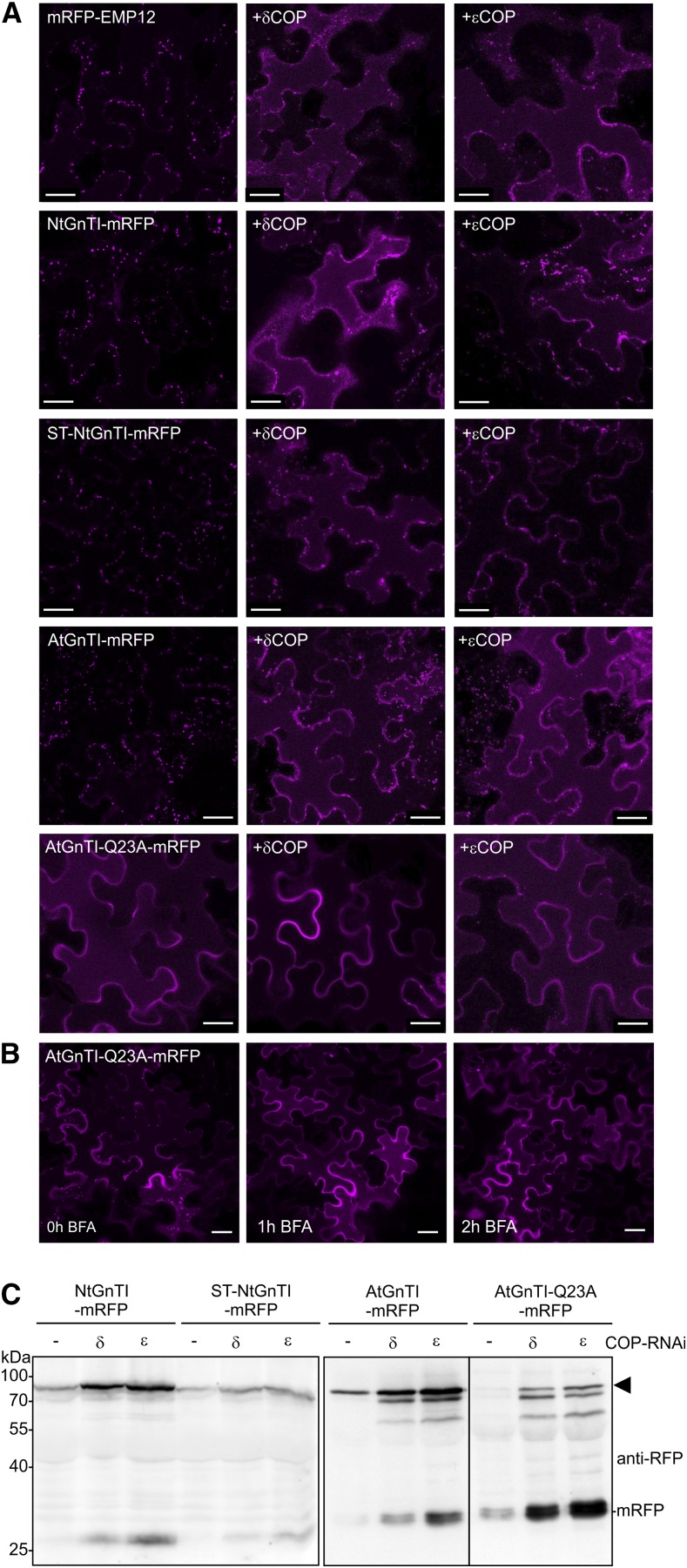
Knockdown of COPI Subunits Leads to GnTI Mislocalization to the Vacuole
To investigate whether COPI subunits are involved in Golgi localization of GnTI, we carried out immunoprecipitation of AtGnTI-mRFP and AtGnTI-Q23A-mRFP and monitored whether endogenous Arabidopsis γCOP is copurified. Immunoblot analysis did not reveal any signal corresponding to γCOP, suggesting that there is no or only a very weak interaction with AtGnTI (Supplemental Fig. S13). Next, we generated RNAi constructs to silence endogenous δCOP and εCOP in N. benthamiana leaves. When the RNAi constructs were transiently coexpressed with the Golgi protein RFP-endomembrane protein12 (RFP-EMP12) that carries a COPI binding site, mistargeting to the vacuole was observed that is in agreement with earlier studies (Gao et al., 2014; Woo et al., 2015). Likewise, NtGnTI-mRFP was mislocalized to the vacuole in cells coinfiltrated with δCOP or εCOP RNAi constructs (Fig. 7A; Supplemental Fig. S13) and the trans-Golgi–resident ST-NtGnTI-mRFP was found extracellularly, as described in Ahn et al. (2015), and in the vacuole (Fig. 7A). Silencing of these COPI subunits targeted AtGnTI-mRFP (or AtGnTI-GFP) to the vacuole, whereas for AtGnTI-mRFP-Q23A, the vacuolar localization was not changed. AtGnTI-mRFP-Q23A was also localized predominately to the vacuole when incubated for 1 or 2 h in brefeldin A (Fig. 7B). Immunoblot analysis revealed an accumulation of the intact mRFP fusion protein upon coexpression of the COPI silencing constructs accompanied by an increase of cleaved mRFP (Fig. 7C). This finding indicates that the Golgi-resident GnTI is not efficiently recycled in the Golgi complex when COPI formation is impaired, leading to vacuolar targeting. Moreover, transient overexpression of AtGnTI-mRFP in N. benthamiana caused an increase in truncated fragments (Supplemental Fig. S13), suggesting a saturation of the GnTI localization mechanism.
Figure 7.
Knockdown of the coatomer subunits δCOP or εCOP results in mislocalization of GnTI. A, Left images show expression in the absence of any RNAi construct. “+δCOP” indicates coinfiltration with the N. benthamiana δCOP RNAi construct. The term “+εCOP” indicates coinfiltration with the N. benthamiana εCOP RNAi construct. mRFP-EMP12 (mRFP-tagged Arabidopsis EMP12 protein lacking the N-terminal luminal domain), NtGnTI-mRFP, ST-NtGnTI-mRFP, AtGnTI-mRFP, and AtGnTI-Q23A-mRFP were observed 48 h after infiltration using confocal microscopy. Scale bars = 25 μm. B, AtGnTI-Q23A-mRFP was treated with brefeldin A (100 μg/mL) for the indicated time. Scale bars = 25 μm. C, Immunoblot analysis of NtGnTI-mRFP, ST-NtGnTI-mRFP, AtGnTI-mRFP, and AtGnTI-Q23A-mRFP. Proteins were coexpressed with the indicated COP RNAi constructs, extracted 72 h after infiltration of N. benthamiana leaves and analyzed with anti-RFP antibodies. An arrowhead marks the intact fusion protein.
DISCUSSION
How can a Distinct Amino Acid Motif within the TMD Mediate Specific Subcellular Localization of a Golgi-Resident Protein?
A large number of secretory proteins are glycosylated and undergo glycan processing reactions in the Golgi. The concentration of processing enzymes in distinct Golgi cisternae is a prerequisite for the sequential and orderly modification of glycans. However, the organization of Golgi-resident enzymes and the mechanism of cargo transport through the Golgi are under debate. In the stable compartment/vesicular transport model, the cargo-processing enzymes remain in their cisternae and cargo is transported from the cis-Golgi to the trans-Golgi by vesicular transport and/or transient tubular connections (Rabouille and Klumperman, 2005; Glick and Luini, 2011). An important feature of the model is the absence of Golgi-resident enzymes in anterograde trafficking carriers and the differential localization of Golgi-resident enzymes is achieved by retention in specific Golgi cisternae. According to this model, the motif in the GnTI TMD constitutes a cis/medial-Golgi retention signal that prevents anterograde transport.
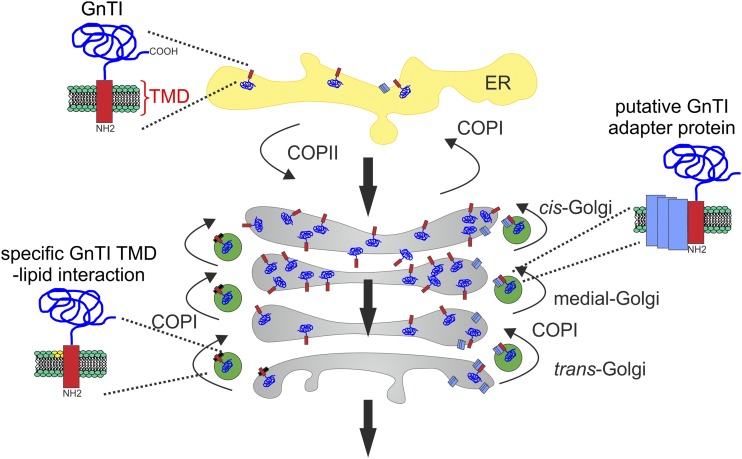
In the cisternal maturation model, cargo stays in the cisternae, which matures from cis-to-trans together with Golgi-resident enzymes. To maintain their gradient-like distribution across the Golgi stack, the cargo-processing enzymes are cycled back from late to earlier Golgi cisternae by COPI vesicles (Villeneuve et al., 2017). The recycling of Golgi processing enzymes transforms biosynthetically inactive cisternae into biosynthetically active cisternae (Donohoe et al., 2013). In the cisternal maturation model, the GnTI TMD motif acts as a signal for incorporation into retrograde transport carriers that mediate recycling of the glycosyltransferase. Even though an interaction with COPI subunits was not found so far, the mislocalization of GnTI to post-Golgi compartments after δCOP and εCOP knockdown can be explained by impaired recycling from the trans-Golgi to an earlier cisternae as proposed by the cisternal maturation model (Fig. 8). The existence of two types of COPI vesicles in plants has been shown by electron tomography (Donohoe et al., 2007). COPIa-type vesicles are located in the vicinity of cis cisternae and likely contribute to Golgi-to-ER transport. COPIb-type vesicles have been found next to medial- and trans-Golgi cisternae. Immunolabeling showed that the cis/medial-Golgi n-glycan processing enzyme MNS1 that generates the n-glycan substrate for GnTI is present in COPIb-type vesicles (Donohoe et al., 2007). Likewise, GnTI may also be incorporated into COPIb-type carriers that cycle from the trans-Golgi to the cis/medial-Golgi. In the absence of recycling, GnTI accumulates in the trans-Golgi from where it is transported to post-Golgi compartments and the vacuole. A comparable recycling mechanism has been proposed for the cis/medial-Golgi–located multiple transmembrane protein AtEMP12 (Gao et al., 2012; Woo et al., 2015). AtEMP12 has a conserved COPI-interacting KXD/E motif in its C-terminal cytoplasmic region. This amino acid sequence is not present in the short cytoplasmic tail of GnTI, and the conserved Gln residue in the exoplasmic half of the transmembrane protein is not accessible for a direct interaction with COPI subunits. Therefore, GnTI may bind via its TMD to an integral membrane protein that serves as an adaptor for incorporation into COPI carriers.
Figure 8.
Proposed working model for GnTI retrieval and its steady-state localization in the cis/medial-Golgi. The model proposes a TMD-motif–based sorting including the retrograde transport from the trans-Golgi with COPI variants (e.g. COPIb; Donohoe et al., 2007). The TMD sequence motif with the conserved Gln is either required for a specific protein-protein interaction with an adaptor protein (indicated on the right side) or for interaction with a specific lipid class or Golgi membrane environment (indicated on the left side) that partitions GnTI into retrograde transport carriers to concentrate the glycosyltransferase in cis/medial-Golgi membranes. The large black arrows indicate cargo that is transported from the ER trough the Golgi apparatus.
The polytopic yeast membrane proteinsER-derived vesicles protein14 andretention in endoplasmic reticulum sorting receptor1 (RER1) are cargo-sorting receptors that recognize TMD features and sort cargo into COPII or COPI carriers (Sato et al., 2001; Herzig et al., 2012; Gomez-Navarro and Miller, 2016). Interestingly, RER1 recognizes polar residues in the TMD of single-TMD proteins such as secretion12 or secretion71 (Sato et al., 2001, 2003) and retrieves them from the Golgi to the ER. Moreover, proper localization of the yeast type-II membrane protein ER α1,2-mannosidaseI depends on the presence of RER1 (Massaad et al., 1999). The TMD of ER α1,2-mannosidase I interacts with RER1, and ER α1,2-mannosidase I is missorted to the vacuole in the absence of a functional RER1. The Arabidopsis RER1 family consists of three members that can complement the mislocalization of cargo proteins in yeast Δrer1, thus suggesting a similar function in plants. However, RER1-interacting proteins and a role for RER1 in intra-Golgi transport have not been described in plants. In addition to ER-derived vesicles protein14 and RER1, EMP family proteins are another group of Golgi-resident integral membrane proteins that may directly interact with GnTI and play a role in its subcellular localization.
Apart from a direct protein-protein interaction and the involvement of an adapter protein, the motif in the GnTI TMD may be crucial for a specific protein-lipid interaction to partition GnTI into a membrane domain competent for recycling to earlier Golgi cisternae (Fig. 8). In mammalian cells, the interaction of a distinct sequence motif in the TMD of p24 with an individual sphingolipid affects the COPI-dependent retrograde transport (Contreras et al., 2012). Moreover, disruption of sphingomyelin homeostasis has led to changes in glycan processing by disturbance of glycosyltransferase organization in distinct Golgi membranes (van Galen et al., 2014). In plants, alterations in the Golgi lipid composition affect protein secretion and Golgi morphology, but the Golgi localization of the trans-Golgi marker ST appears unaltered (Melser et al., 2010).
The function of GnTI in n-glycan processing as well as its subcellular localization are conserved in mammals and plants (von Schaewen et al., 1993; Gomez and Chrispeels, 1994; Bakker et al., 1999; Strasser et al., 1999) and there is evidence for cycling of mammalian GnTI (Hoe et al., 1995). A recently identified COPI-binding motif in mammalian glycosyltransferases is not present in the cytoplasmic tail of human GnTI and, in contrast to other cis/medial-Golgi enzymes, no direct binding of GnTI to COPI subunits was found (Liu et al., 2018). GOLPH3, the mammalian VPS74 homolog, is involved in the incorporation of certain glycosyltransferases into COPI vesicles (Eckert et al., 2014) but its absence had no effect on GnTI localization in human cells (Liu et al., 2018). A role for the TMD of mammalian GnTI in Golgi localization has also been described in other studies (Burke et al., 1992, 1994; Tang et al., 1992; Nilsson et al., 1996) and the TMD from mammalian GnTIs share a sequence motif with a highly conserved Asn residue at a similar position. Whether this residue or other features of the TMD are involved in intra-Golgi localization of mammalian GnTI remains to be shown. A general role of the TMD in Golgi retention of type-II membrane proteins has been proposed for yeast and mammals (Quiroga et al., 2013), and individual amino acids including polar residues within the TMD contribute to Golgi localization of mammalian proteins (Aoki et al., 1992; Machamer et al., 1993), but a specific receptor or lipid interaction has not been described.
In summary, our data show that a transmembrane-motif–based sorting mechanism mediates the steady-state localization of GnTI in plants. Our findings are fully consistent with the fact that GnTI homomer formation plays no or only a minor role in cis/medial Golgi localization (Schoberer et al., 2013, 2014). The discovered mechanism that is functional in Arabidopsis as well as in N. benthamiana is distinct from the earlier proposed bilayer thickness-mediated retention, the kin recognition, and the more recently recognized cytoplasmic tail-dependent sorting (Tu and Banfield, 2010). Our model proposes that a conserved polar amino acid in the TMD promotes the interaction with a yet-unknown adaptor protein/complex or the segregation into a distinct lipid domain. Impaired recycling in the absence of the specific interaction results in GnTI exit from the Golgi and trafficking to post-Golgi compartments, including the vacuole where GnTI is eventually degraded. This trafficking route may highlight the default degradation pathway for glycosyltransferases that are not needed anymore in a plant cell.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana; ecotype Col-0) and mutant plants were grown at 22°C under long-day conditions (16-h-light/8-h-dark photoperiod) on soil or on 0.5× Murashige and Skoog medium containing 1% (w/v) Suc and 0.8% (w/v) agar. The mns1 mns2 mns3 (Liebminger et al., 2009), gntI (Schoberer et al., 2014), and cgl1 (von Schaewen et al., 1993; Strasser et al., 2005) mutants were available from previous studies. For the measurement of the root length, Arabidopsis seedlings were grown for 10 d on 0.5× Murashige and Skoog medium supplemented with 4% (w/v) Suc. For the inhibitor treatments, Arabidopsis seedlings were incubated for 17 h in liquid Murashige and Skoog medium supplemented with 1 μM of concanamycin A (Sigma-Aldrich), 20 μM of MG132 (Sigma-Aldrich), 100 μg/mL of CHX (Sigma-Aldrich), or 20 μM of kifunensine (Santa Cruz Biotechnology). Nicotiana benthamiana were grown under long-day conditions at 24°C.
Cloning of Constructs
DNA fragments coding for the chimeric GnTI/ST-, ST/GnTI-, and STFIYIQ-CTS regions were obtained by custom DNA synthesis (GeneArt; Thermo Fisher Scientific) and ligated into XbaI/BamHI sites of p20-Fc (CaMV35S promoter, used for colocalization analysis; Supplemental Fig. S2; Schoberer et al., 2009), pF (CaMV35S promoter, GALT fusions containing the catalytic domain of human GALT, used for the n-glycan processing assay; Schoberer et al., 2014), p46 (UBQ10 promoter, expression of proteins containing a CTS region fused to the catalytic domain of NtGnTI and GFP; Schoberer et al., 2014), p49 (like p46 but with a C-terminal mRFP tag), p57 (386-bp weak AtGnTI promoter, complementation of gntI plants with AtGnTI catalytic domain fused to GFP; Schoberer et al., 2014), p66 (UBQ10 promoter, expression of proteins containing a CTS region fused to the catalytic domain of AtGnTI and GFP), p67 (1,000-bp AtGnTI promoter, complementation of gntI plants with AtGnTI catalytic domain fused to GFP), p68 (like p67 but with a C-terminal mRFP tag), and p69 (like p66 but with a C-terminal mRFP tag). Plant expression vector p67 was generated by insertion of the 1,000-bp AtGnTI promoter region into the HindIII/XbaI site of p57. The 1,000-bp AtGnTI promoter region was amplified from Arabidopsis genomic DNA by PCR using AthGnT33/AthGnTI_13R (Supplemental Table S2). p68 was generated by replacing GFP in vector p67 with mRFP. p66 and p69 were generated by replacement of the AtGnTI promoter with the UBQ10 promoter.
The constructs for expression of the glycoprotein reporter (monoclonal antibody; Strasser et al., 2009), ST-mRFP, ST-GFP, NtGnTI-CTS-GFP (Boevink et al., 1998; Schoberer et al., 2009), mRFP-AtCASP (Schoberer et al., 2010), NtGnTI-GFP, NtGnTI-mRFP, ST-NtGnTI-GFP, ST-NtGnTI-mRFP (Schoberer et al., 2014), aleu-GFP (Shin et al., 2017), mRFP-EMP12 (Shin et al., 2018), mRFP-SYP61 (Sanderfoot et al., 2001), GFP-RABF2b (Kotzer et al., 2004), and GFP-RABF1 (Ueda et al., 2001) were all available from previous studies. p48-MUR3 was generated by PCR amplification from Col-0 cDNA of the MUR3 (AT2G20370) coding region using primers KAM1_1F/KAM1_3R. The PCR product was XbaI/BglII-digested and cloned into p48 (Hüttner et al., 2014). For GFP-ATG8e expression, the coding region for Arabidopsis ATG8e (AT2G45170) was amplified by PCR from Col-0 cDNA using primers ATG8e_1F/ATG8e_2R. The PCR product was XbaI/BamHI-digested and cloned into XbaI/BamHI-digested vector p37 (CaMV35S promoter). For the generation of p37, the GFP coding region lacking the stop codon was PCR-amplified from the vector p20F using primers GFP-14F/GFP-15R. The PCR product was SpeI/XbaI-digested and ligated into XbaI-linearized pPT2m (Strasser et al., 2005) resulting in the N-terminal GFP expression vector p37.
NtGnTI and AtGnTI CTS regions with mutated single amino acids were generated by site-directed mutagenesis using the QuikChange-site–directed mutagenesis protocol (Stratagene) and cloned into the XbaI/BamHI sites of the used expression vectors. The expression construct p87 (MNS1 promoter, MNS1 catalytic domain fused to GFP) for the chimeric MNS1 fusion proteins was described in Veit et al. (2018). For expression of AtGnTI and AtGnTI-Q23A CTS regions, the corresponding DNA was amplified with primers AthGnTI_14F/AthGnTI_16R and ligated into XbaI/BamHI-digested p87.
Liquid-Chromatography Electrospray Ionization-Mass Spectrometry Analysis
Five-week–old N. benthamiana plants were used for Agrobacterium tumefaciens-mediated transient expression of indicated constructs using the agroinfiltration technique as described in Schoberer et al. (2009). The glycoprotein reporter was purified by affinity chromatography, subjected to SDS-PAGE and Coomassie Brilliant Blue staining (Schoberer et al., 2014). The purified protein band was excised from the gel, destained, carbamidomethylated, in-gel-trypsin–digested, and analyzed by liquid-chromatography electrospray ionization-mass spectrometry (LC-ESI-MS; Stadlmann et al., 2008; Schoberer et al., 2009). A detailed explanation of n-glycan abbreviations and nomenclature can be found at http://www.proglycan.com.
Complementation of Arabidopsis Mutants
Arabidopsis gntI, cgl1, and mns1 mns2 mns3 mutants were transformed with different constructs by floral dipping and selected on hygromycin (Strasser et al., 2004). The presence of the construct was confirmed by PCR with specific primers. Proteins extracted from leaves of 5-week–old plants were subjected to SDS-PAGE under reducing conditions. Glycoproteins with processed n-glycans were analyzed by immunoblotting with anti-horseradish peroxidase antibodies (anti-HRP; Sigma-Aldrich) that bind to complex, truncated, and hybrid n-glycans carrying β1,2-Xyl and/or core α1,3-Fuc residues (Strasser et al., 2004; Jin et al., 2008; Kaulfürst-Soboll et al., 2011). MALDI-MS analysis was carried out from 500 mg of leaves as described in Strasser et al. (2004).
Confocal Imaging of Fluorescent Protein Fusions
Leaves of 5-week–old N. benthamiana plants were infiltrated with agrobacterium suspensions carrying the protein(s) of interest with optical densities (OD600) from 0.05 to 0.2. Confocal images were acquired 2–4 d post infiltration (dpi) on an upright SP5 II Confocal Microscope (Leica) using the LAS AF software system (Leica). GFP and mRFP were excited with the 488-nm and 561-nm laser line, respectively, and detected at 500–530 nm and 600–630 nm, respectively. Dual-color image acquisition of cells expressing both GFP and mRFP was performed simultaneously. Postacquisition image processing was performed in Adobe Photoshop CS5.
For colocalization analyses of coexpressed fluorescent protein fusions, images of latrunculin-B–treated cells expressing the indicated NtGnTI and ST-CTS constructs together with the cis/medial-Golgi protein mRFP-AtCASP (Schoberer et al., 2010) or the trans-Golgi marker ST-mRFP (Boevink et al., 1998) were acquired 2 dpi under nonsaturating conditions using a 512 × 512 image format, zoom factor 3, and a ×100/1.40 NA oil immersion objective of the Leica SP5. The pinhole was set to 1 Airy unit, and background noise was reduced by 4–8-times line averaging. Only cells with comparable GFP and mRFP fluorescence levels were considered for analysis. The images obtained were used for colocalization analysis using the Pearson’s correlation coefficient. Calculations were made on 9–15 confocal images per coexpressed combination using the software ImageJ’s 1.46 m plug-in JACOP (Bolte and Cordelières, 2006). Statistical analyses were performed using a two-tailed Student’s t test for the comparison of two samples, assuming equal variances. FRET-FLIM data acquisition and analysis was carried out as described in Schoberer et al. (2014).
Coimmunoprecipitation and Immunoblotting
For analysis of AtGnTI-Q23A and AtGnTI interaction, AtGnTI-Q23A-mRFP (p69 vector), or AtGnTI-mRFP (p69 vector) was coexpressed with AtGnTI-GFP (p66 vector) by agroinfiltration of N. benthamiana leaves. Two dpi, the GFP-tagged protein was purified using GFP-Trap-A beads (ChromoTek) and copurified protein was analyzed by immunoblotting with anti-mRFP antibodies. Next to BRCA1 GENE1 (NBR1) was detected with anti-NBR1 (Agrisera), and antibodies against actin (Agrisera) and α-tubulin (Sigma-Aldrich) were used as loading controls.
RT-qPCR Analysis
RNA was extracted from 18-d–old Arabidopsis seedlings using the SV Total RNA Isolation System (Promega) and the iScript cDNA Synthesis Kit (Bio-Rad). An aliquot of the cDNA was used for the PCR amplification using iQ SYBR Green Supermix (Bio-Rad) with an iCycler (Bio-Rad) as described in detail in Hüttner et al. (2014). Expression of GFP-fusion proteins was monitored using GFP-specific primers GFP-Q1F and GFP-Q2R and normalized to the UBQ5 transcript expression monitored with primers UBQ5_Q1F/UBQ5_Q2R.
Knockdown of N. benthamiana Coatomer Subunits
To knockdown the expression of the N. benthamiana εCOP and δCOP subunits, RNAi constructs were generated as described in Shin et al. (2017). A synthetic DNA fragment consisting of intron 2 from Arabidopsis β1,2-xylosyltransferase and an antisense DNA fragment corresponding to the coding sequence for amino acids 35–186 of εCOP or 306–473 of δCOP were obtained by GeneArt gene synthesis. The vector with the synthetic DNA was used as a template for PCR with primers Nb-eCOP-1F and Nb-eCOP-2R for εCOP and Nb-dCOP-1F and Nb-dCOP-2R for δCOP. The “sense” PCR product was digested with XbaI/KpnI and cloned into the synthetic DNA containing vector to generate a sense–intron–antisense hairpin construct. The sense–intron–antisense sequence was subsequently excised by XbaI/BamHI digestion and ligated into XbaI/BamHI-digested plant expression vector pPT2m. The pPT2-NbεCOP-RNAi and pPT2-NbδCOP-RNAi vectors were transformed into agrobacteria and transiently expressed by agroinfiltration in N. benthamiana leaves together with the fluorescent-tagged proteins.
Accession Numbers
Sequence data from this article can be found in the GenBank database under the following accession numbers: M18769 (ST) and AJ295993 (NtGnTI); in the Arabidopsis Information Resource under the following accession numbers: AT4G38240 (AtGnTI) and AT1G51590 (MNS1); and in the N. benthamiana draft genome sequence v1.0.1 from the Sol Genomics Network: Niben101Scf03249g01008 (εCOP) and Niben101Scf09552g01014 (δCOP).
Supplemental Data
The following supplemental information is available.
Supplemental Figure S1. Amino acid sequence alignment of the N-terminal sequence (including the TMD) from 74 putative plant GnTI sequences.
Supplemental Figure S2. CTS regions and used expression vectors.
Supplemental Figure S3. Subcellular localization of fusion proteins containing a chimeric or mutated CTS region.
Supplemental Figure S4. Schematic illustration of the n-glycan processing assay.
Supplemental Figure S5. LC-ESI-MS analysis of the glycopeptide derived from a secreted glycoreporter (IgG antibody) coexpressed with ST/GnTI-GALT, GnTI/ST-GALT, or STFIYIQ-GALT.
Supplemental Figure S6. Complementation of the Arabidopsis gntI n-glycan processing defect.
Supplemental Figure S7. Complementation of Arabidopsis gntI and cgl1 plants.
Supplemental Figure S8. MALDI-TOF spectra of gntI expressing the indicated constructs under the control of a weak AtGnTI promoter (386 bp).
Supplemental Figure S9. Colocalization of mutated AtGnTI with the trans-Golgi marker ST-GFPglyc.
Supplemental Figure S10. Comparison of the TMD sequence composition.
Supplemental Figure S11. AtGnTI variants with other conserved amino acids mutated reside in the Golgi and are functional.
Supplemental Figure S12. Phenotypic comparison of complemented mns1 mns2 mns3 seedlings.
Supplemental Figure S13. Knockdown of the coatomer subunits δCOP or εCOP results in mislocalization of Golgi-resident GnTI.
Supplemental Table S1. Statistical analyses of colocalization data.
Supplemental Table S2. List of used primers.
Acknowledgments
We thank Martina Dicker, Josephine Grass, Julia König, Daniel Maresch, Karin Polacsek, and Yun-ji Shin (all from BOKU-University of Natural Resources and Life Sciences, Vienna) for technical assistance and help with n-glycan and glycopeptide analysis. Verena Kriechbaumer (Oxford Brookes University) is acknowledged for supplying plants used at the Central Laser Facility. We thank BOKU-Vienna Institute of Technology Imaging Center for access and expertise.
Footnotes
This work was supported by the Austrian Science Fund (grants no. P23906-B20 and P28218-B22 to R.S., and T655-B20 to J.S.), the Mizutani Foundation for Glycosciences (to R.S.), and the Science and Technology Facilities Council (grant no. 12130006 to Chris Hawes, Oxford Brookes University, for access to the Central Laser Facility).
Articles can be viewed without a subscription.
References
- Ahn HK, Kang YW, Lim HM, Hwang I, Pai HS (2015) Physiological functions of the COPI complex in higher plants. Mol Cells 38: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki D, Lee N, Yamaguchi N, Dubois C, Fukuda MN (1992) Golgi retention of a trans-Golgi membrane protein, galactosyltransferase, requires cysteine and histidine residues within the membrane-anchoring domain. Proc Natl Acad Sci USA 89: 4319–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker H, Lommen A, Jordi W, Stiekema W, Bosch D (1999) An Arabidopsis thaliana cDNA complements the n-acetylglucosaminyltransferase I deficiency of CHO Lec1 cells. Biochem Biophys Res Commun 261: 829–832 [DOI] [PubMed] [Google Scholar]
- Boevink P, Oparka K, Santa Cruz S, Martin B, Betteridge A, Hawes C (1998) Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J 15: 441–447 [DOI] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232 [DOI] [PubMed] [Google Scholar]
- Burke J, Pettitt JM, Schachter H, Sarkar M, Gleeson PA (1992) The transmembrane and flanking sequences of beta 1,2-n-acetylglucosaminyltransferase I specify medial-Golgi localization. J Biol Chem 267: 24433–24440 [PubMed] [Google Scholar]
- Burke J, Pettitt JM, Humphris D, Gleeson PA (1994) Medial-Golgi retention of n-acetylglucosaminyltransferase I. Contribution from all domains of the enzyme. J Biol Chem 269: 12049–12059 [PubMed] [Google Scholar]
- Chamberlain KL, Marshall RS, Jolliffe NA, Frigerio L, Ceriotti A, Lord JM, Roberts LM (2008) Ricin B chain targeted to the endoplasmic reticulum of tobacco protoplasts is degraded by a CDC48- and vacuole-independent mechanism. J Biol Chem 283: 33276–33286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier L, Bernard S, Ramdani Y, Lamour R, Bardor M, Lerouge P, Follet-Gueye ML, Driouich A (2010) Subcompartment localization of the side chain xyloglucan-synthesizing enzymes within Golgi stacks of tobacco suspension-cultured cells. Plant J 64: 977–989 [DOI] [PubMed] [Google Scholar]
- Contento AL, Xiong Y, Bassham DC (2005) Visualization of autophagy in Arabidopsis using the fluorescent dye monodansylcadaverine and a GFP-AtATG8e fusion protein. Plant J 42: 598–608 [DOI] [PubMed] [Google Scholar]
- Contreras FX, Ernst AM, Haberkant P, Björkholm P, Lindahl E, Gönen B, Tischer C, Elofsson A, von Heijne G, Thiele C, et al. (2012) Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature 481: 525–529 [DOI] [PubMed] [Google Scholar]
- Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K (2006) Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe BS, Kang BH, Staehelin LA (2007) Identification and characterization of COPIa- and COPIb-type vesicle classes associated with plant and algal Golgi. Proc Natl Acad Sci USA 104: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe BS, Kang BH, Gerl MJ, Gergely ZR, McMichael CM, Bednarek SY, Staehelin LA (2013) Cis-Golgi cisternal assembly and biosynthetic activation occur sequentially in plants and algae. Traffic 14: 551–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunphy WG, Rothman JE (1983) Compartmentation of asparagine-linked oligosaccharide processing in the Golgi apparatus. J Cell Biol 97: 270–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert ES, Reckmann I, Hellwig A, Röhling S, El-Battari A, Wieland FT, Popoff V (2014) Golgi phosphoprotein 3 triggers signal-mediated incorporation of glycosyltransferases into coatomer-coated (COPI) vesicles. J Biol Chem 289: 31319–31329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanata WI, Lee KH, Son BH, Yoo JY, Harmoko R, Ko KS, Ramasamy NK, Kim KH, Oh DB, Jung HS, et al. (2013) n-glycan maturation is crucial for cytokinin-mediated development and cellulose synthesis in Oryza sativa. Plant J 73: 966–979 [DOI] [PubMed] [Google Scholar]
- Fisher P, Ungar D (2016) Bridging the gap between glycosylation and vesicle traffic. Front Cell Dev Biol 4: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Yu CK, Qu S, San MW, Li KY, Lo SW, Jiang L (2012) The Golgi-localized Arabidopsis endomembrane protein12 contains both endoplasmic reticulum export and Golgi retention signals at its C terminus. Plant Cell 24: 2086–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Cai Y, Wang Y, Kang BH, Aniento F, Robinson DG, Jiang L (2014) Retention mechanisms for ER and Golgi membrane proteins. Trends Plant Sci 19: 508–515 [DOI] [PubMed] [Google Scholar]
- Glick BS, Luini A (2011) Models for Golgi traffic: A critical assessment. Cold Spring Harb Perspect Biol 3: a005215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez L, Chrispeels MJ (1994) Complementation of an Arabidopsis thaliana mutant that lacks complex asparagine-linked glycans with the human cDNA encoding n-acetylglucosaminyltransferase I. Proc Natl Acad Sci USA 91: 1829–1833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Navarro N, Miller E (2016) Protein sorting at the ER-Golgi interface. J Cell Biol 215: 769–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A, Aebi M (2001) Intracellular functions of n-linked glycans. Science 291: 2364–2369 [DOI] [PubMed] [Google Scholar]
- Herzig Y, Sharpe HJ, Elbaz Y, Munro S, Schuldiner M (2012) A systematic approach to pair secretory cargo receptors with their cargo suggests a mechanism for cargo selection by Erv14. PLoS Biol 10: e1001329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe MH, Slusarewicz P, Misteli T, Watson R, Warren G (1995) Evidence for recycling of the resident medial/trans Golgi enzyme, n-acetylglucosaminyltransferase I, in ldlD cells. J Biol Chem 270: 25057–25063 [DOI] [PubMed] [Google Scholar]
- Humair D, Hernández Felipe D, Neuhaus JM, Paris N (2001) Demonstration in yeast of the function of BP-80, a putative plant vacuolar sorting receptor. Plant Cell 13: 781–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttner S, Veit C, Vavra U, Schoberer J, Liebminger E, Maresch D, Grass J, Altmann F, Mach L, Strasser R (2014) Arabidopsis class I α-mannosidases MNS4 and MNS5 are involved in endoplasmic reticulum-associated degradation of misfolded glycoproteins. Plant Cell 26: 1712–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioffe E, Stanley P (1994) Mice lacking n-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid n-linked carbohydrates. Proc Natl Acad Sci USA 91: 728–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Uemura T, Nakano A (2014) Formation and maintenance of the Golgi apparatus in plant cells. Int Rev Cell Mol Biol 310: 221–287 [DOI] [PubMed] [Google Scholar]
- Jin C, Altmann F, Strasser R, Mach L, Schähs M, Kunert R, Rademacher T, Glössl J, Steinkellner H (2008) A plant-derived human monoclonal antibody induces an anti-carbohydrate immune response in rabbits. Glycobiology 18: 235–241 [DOI] [PubMed] [Google Scholar]
- Kang JS, Frank J, Kang CH, Kajiura H, Vikram M, Ueda A, Kim S, Bahk JD, Triplett B, Fujiyama K, et al. (2008) Salt tolerance of Arabidopsis thaliana requires maturation of n-glycosylated proteins in the Golgi apparatus. Proc Natl Acad Sci USA 105: 5933–5938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulfürst-Soboll H, Rips S, Koiwa H, Kajiura H, Fujiyama K, von Schaewen A (2011) Reduced immunogenicity of Arabidopsis hgl1 mutant n-glycans caused by altered accessibility of xylose and core fucose epitopes. J Biol Chem 286: 22955–22964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotzer AM, Brandizzi F, Neumann U, Paris N, Moore I, Hawes C (2004) AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J Cell Sci 117: 6377–6389 [DOI] [PubMed] [Google Scholar]
- Liebminger E, Hüttner S, Vavra U, Fischl R, Schoberer J, Grass J, Blaukopf C, Seifert GJ, Altmann F, Mach L, et al. (2009) Class I alpha-mannosidases are required for n-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21: 3850–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Doray B, Kornfeld S (2018) Recycling of Golgi glycosyltransferases requires direct binding to coatomer. Proc Natl Acad Sci USA 115: 8984–8989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE. (1991) Golgi retention signals: Do membranes hold the key? Trends Cell Biol 1: 141–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machamer CE, Grim MG, Esquela A, Chung SW, Rolls M, Ryan K, Swift AM (1993) Retention of a cis Golgi protein requires polar residues on one face of a predicted alpha-helix in the transmembrane domain. Mol Biol Cell 4: 695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaad MJ, Franzusoff A, Herscovics A (1999) The processing alpha1,2-mannosidase of Saccharomyces cerevisiae depends on Rer1p for its localization in the endoplasmic reticulum. Eur J Cell Biol 78: 435–440 [DOI] [PubMed] [Google Scholar]
- Melser S, Batailler B, Peypelut M, Poujol C, Bellec Y, Wattelet-Boyer V, Maneta-Peyret L, Faure JD, Moreau P (2010) Glucosylceramide biosynthesis is involved in Golgi morphology and protein secretion in plant cells. Traffic 11: 479–490 [DOI] [PubMed] [Google Scholar]
- Munro S. (1995) An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J 14: 4695–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Slusarewicz P, Hoe MH, Warren G (1993) Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett 330: 1–4 [DOI] [PubMed] [Google Scholar]
- Nilsson T, Rabouille C, Hui N, Watson R, Warren G (1996) The role of the membrane-spanning domain and stalk region of n-acetylglucosaminyltransferase I in retention, kin recognition and structural maintenance of the Golgi apparatus in HeLa cells. J Cell Sci 109: 1975–1989 [DOI] [PubMed] [Google Scholar]
- Osterrieder A, Hummel E, Carvalho CM, Hawes C (2010) Golgi membrane dynamics after induction of a dominant-negative mutant Sar1 GTPase in tobacco. J Exp Bot 61: 405–422 [DOI] [PubMed] [Google Scholar]
- Patterson GH, Hirschberg K, Polishchuk RS, Gerlich D, Phair RD, Lippincott-Schwartz J (2008) Transport through the Golgi apparatus by rapid partitioning within a two-phase membrane system. Cell 133: 1055–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen CT, Loke I, Lorentzen A, Wolf S, Kamble M, Kristensen SK, Munch D, Radutoiu S, Spillner E, Roepstorff P, et al. (2017) N-glycan maturation mutants in Lotus japonicus for basic and applied glycoprotein research. Plant J 91: 394–407 [DOI] [PubMed] [Google Scholar]
- Petrosyan A, Ali MF, Cheng PW (2015) Keratin 1 plays a critical role in Golgi localization of core 2 n-acetylglucosaminyltransferase M via interaction with its cytoplasmic tail. J Biol Chem 290: 6256–6269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga R, Trenchi A, González Montoro A, Valdez Taubas J, Maccioni HJ (2013) Short transmembrane domains with high-volume exoplasmic halves determine retention of Type II membrane proteins in the Golgi complex. J Cell Sci 126: 5344–5349 [DOI] [PubMed] [Google Scholar]
- Rabouille C, Klumperman J (2005) Opinion: The maturing role of COPI vesicles in intra-Golgi transport. Nat Rev Mol Cell Biol 6: 812–817 [DOI] [PubMed] [Google Scholar]
- Rabouille C, Hui N, Hunte F, Kieckbusch R, Berger EG, Warren G, Nilsson T (1995) Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J Cell Sci 108: 1617–1627 [DOI] [PubMed] [Google Scholar]
- Reichardt I, Stierhof YD, Mayer U, Richter S, Schwarz H, Schumacher K, Jürgens G (2007) Plant cytokinesis requires de novo secretory trafficking but not endocytosis. Curr Biol 17: 2047–2053 [DOI] [PubMed] [Google Scholar]
- Runions J, Brach T, Kühner S, Hawes C (2006) Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J Exp Bot 57: 43–50 [DOI] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C, Nebenführ A, Boulaflous A, Follet-Gueye ML, Plasson C, Hawes C, Driouich A, Faye L, Gomord V (2006) Plant n-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18: 3182–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV (2001) Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell 12: 3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A (2001) Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J Cell Biol 152: 935–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sato M, Nakano A (2003) Rer1p, a retrieval receptor for ER membrane proteins, recognizes transmembrane domains in multiple modes. Mol Biol Cell 14: 3605–3616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz KR, Liu J, Li S, Setty TG, Wood CS, Burd CG, Ferguson KM (2008) Golgi localization of glycosyltransferases requires a Vps74p oligomer. Dev Cell 14: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Vavra U, Stadlmann J, Hawes C, Mach L, Steinkellner H, Strasser R (2009) Arginine/lysine residues in the cytoplasmic tail promote ER export of plant glycosylation enzymes. Traffic 10: 101–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Runions J, Steinkellner H, Strasser R, Hawes C, Osterrieder A (2010) Sequential depletion and acquisition of proteins during Golgi stack disassembly and reformation. Traffic 11: 1429–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Liebminger E, Botchway SW, Strasser R, Hawes C (2013) Time-resolved fluorescence imaging reveals differential interactions of n-glycan processing enzymes across the Golgi stack in planta. Plant Physiol 161: 1737–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Liebminger E, Vavra U, Veit C, Castilho A, Dicker M, Maresch D, Altmann F, Hawes C, Botchway SW, et al. (2014) The transmembrane domain of n-acetylglucosaminyltransferase I is the key determinant for its Golgi subcompartmentation. Plant J 80: 809–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe HJ, Stevens TJ, Munro S (2010) A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142: 158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YJ, Castilho A, Dicker M, Sádio F, Vavra U, Grünwald-Gruber C, Kwon TH, Altmann F, Steinkellner H, Strasser R (2017) Reduced paucimannosidic n-glycan formation by suppression of a specific β-hexosaminidase from Nicotiana benthamiana. Plant Biotechnol J 15: 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YJ, Vavra U, Veit C, Strasser R (2018) The glycan-dependent ERAD machinery degrades topologically diverse misfolded proteins. Plant J 94: 246–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F (2008) Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics 8: 2858–2871 [DOI] [PubMed] [Google Scholar]
- Strasser R. (2016) Plant protein glycosylation. Glycobiology 26: 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Mucha J, Schwihla H, Altmann F, Glössl J, Steinkellner H (1999) Molecular cloning and characterization of cDNA coding for beta1, 2n-acetylglucosaminyltransferase I (GlcNAc-TI) from Nicotiana tabacum. Glycobiology 9: 779–785 [DOI] [PubMed] [Google Scholar]
- Strasser R, Altmann F, Mach L, Glössl J, Steinkellner H (2004) Generation of Arabidopsis thaliana plants with complex n-glycans lacking beta1,2-linked xylose and core alpha1,3-linked fucose. FEBS Lett 561: 132–136 [DOI] [PubMed] [Google Scholar]
- Strasser R, Stadlmann J, Svoboda B, Altmann F, Glössl J, Mach L (2005) Molecular basis of n-acetylglucosaminyltransferase I deficiency in Arabidopsis thaliana plants lacking complex n-glycans. Biochem J 387: 385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Castilho A, Stadlmann J, Kunert R, Quendler H, Gattinger P, Jez J, Rademacher T, Altmann F, Mach L, et al. (2009) Improved virus neutralization by plant-produced anti-HIV antibodies with a homogeneous beta1,4-galactosylated n-glycan profile. J Biol Chem 284: 20479–20485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang BL, Wong SH, Low SH, Hong W (1992) The transmembrane domain of n-glucosaminyltransferase I contains a Golgi retention signal. J Biol Chem 267: 10122–10126 [PubMed] [Google Scholar]
- Tu L, Banfield DK (2010) Localization of Golgi-resident glycosyltransferases. Cell Mol Life Sci 67: 29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L, Tai WC, Chen L, Banfield DK (2008) Signal-mediated dynamic retention of glycosyltransferases in the Golgi. Science 321: 404–407 [DOI] [PubMed] [Google Scholar]
- Ueda T, Yamaguchi M, Uchimiya H, Nakano A (2001) Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J 20: 4730–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uliana AS, Giraudo CG, Maccioni HJ (2006) Cytoplasmic tails of SialT2 and GalNAcT impose their respective proximal and distal Golgi localization. Traffic 7: 604–612 [DOI] [PubMed] [Google Scholar]
- van Galen J, Campelo F, Martínez-Alonso E, Scarpa M, Martínez-Menárguez JÁ, Malhotra V (2014) Sphingomyelin homeostasis is required to form functional enzymatic domains at the trans-Golgi network. J Cell Biol 206: 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit C, König J, Altmann F, Strasser R (2018) Processing of the terminal alpha-1,2-linked mannose residues from oligomannosidic n-glycans is critical for proper root growth. Front Plant Sci 9: 1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve J, Duran J, Scarpa M, Bassaganyas L, Van Galen J, Malhotra V (2017) Golgi enzymes do not cycle through the endoplasmic reticulum during protein secretion or mitosis. Mol Biol Cell 28: 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schaewen A, Sturm A, O’Neill J, Chrispeels MJ (1993) Isolation of a mutant Arabidopsis plant that lacks n-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex n-linked glycans. Plant Physiol 102: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CH, Gao C, Yu P, Tu L, Meng Z, Banfield DK, Yao X, Jiang L (2015) Conserved function of the lysine-based KXD/E motif in Golgi retention for endomembrane proteins among different organisms. Mol Biol Cell 26: 4280–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]