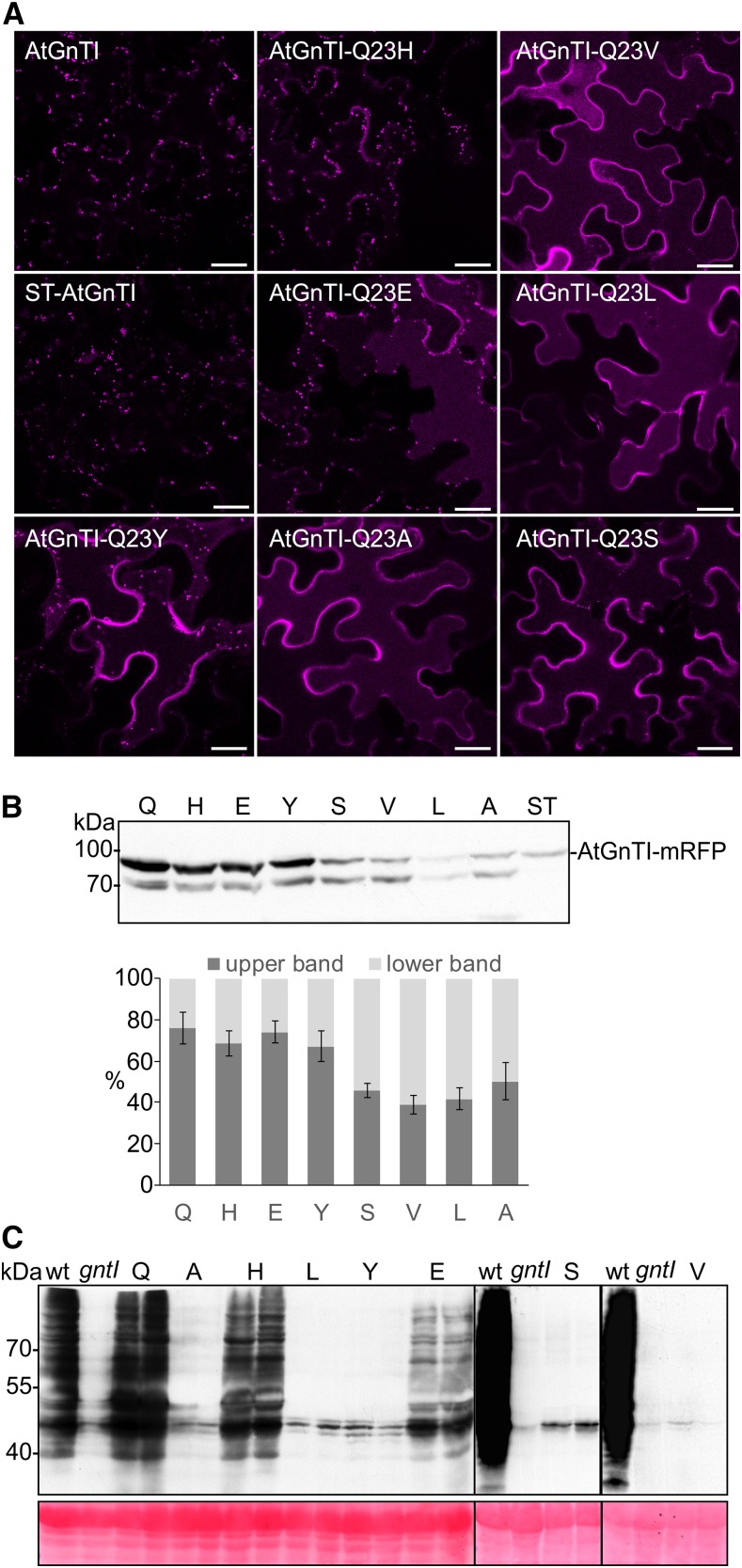
Figure 5.
AtGnTI variants with an altered amino acid at position 23 are mistargeted in plants and do not complement the gntI n-glycan processing defect. A, Confocal microscopy analysis in N. benthamiana leaf epidermal cells of transiently expressed AtGnTI-Q23-mRFP mutants. Confocal images were acquired 48 h after infiltration. B, Transiently expressed AtGnTI-Q23-mRFP mutants analyzed by immunoblotting using an anti-mRFP antibody. The quantitation shows the relative amount of the upper (intact AtGnTI-mRFP) and the lower (AtGnTI-mRFP degradation product) bands. Values are means ± sd (n = 4 independent experiments. The single amino acid labeling indicates the substituted amino acid at position 23. C, Immunoblot analysis of transgenic gntI plants expressing the different AtGnTI-Q23-mRFP variants under the control of the endogenous AtGnTI promoter. Protein extracts from leaves were analyzed with antibodies against complex plant n-glycans (anti-HRP). Two independent lines are shown for each AtGnTI-Q23-mRFP variant, Col-0 (wild type), and gntI protein extracts were included as controls. Ponceau S staining of the membrane was used as a loading control.