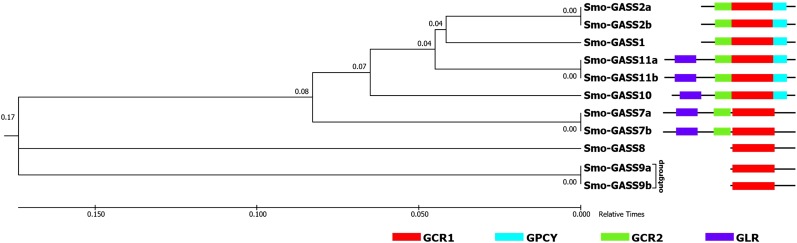
Figure 4.
The reconstructed TimeTree was used to predict the evolution of S. moellendorffii GASTs containing conserved motifs in a relative time frame. A TimeTree was reconstructed from S. moellendorffii GAST orthologs using the RelTime method inferred from neighbour joining in MEGA7 (Kumar et al., 2016). The TimeTree indicated the evolution of GASTs according to the presence of conserved motifs. Smo-GASS9a and Smo-GASS9b were taken as the outgroup and the distance was calculated in a relative time of 1. On the right, the GAST sequences are shown with their respective motifs arranged according to their evolution. The Smo-GASS sequences and length of the motifs do not represent actual scale.