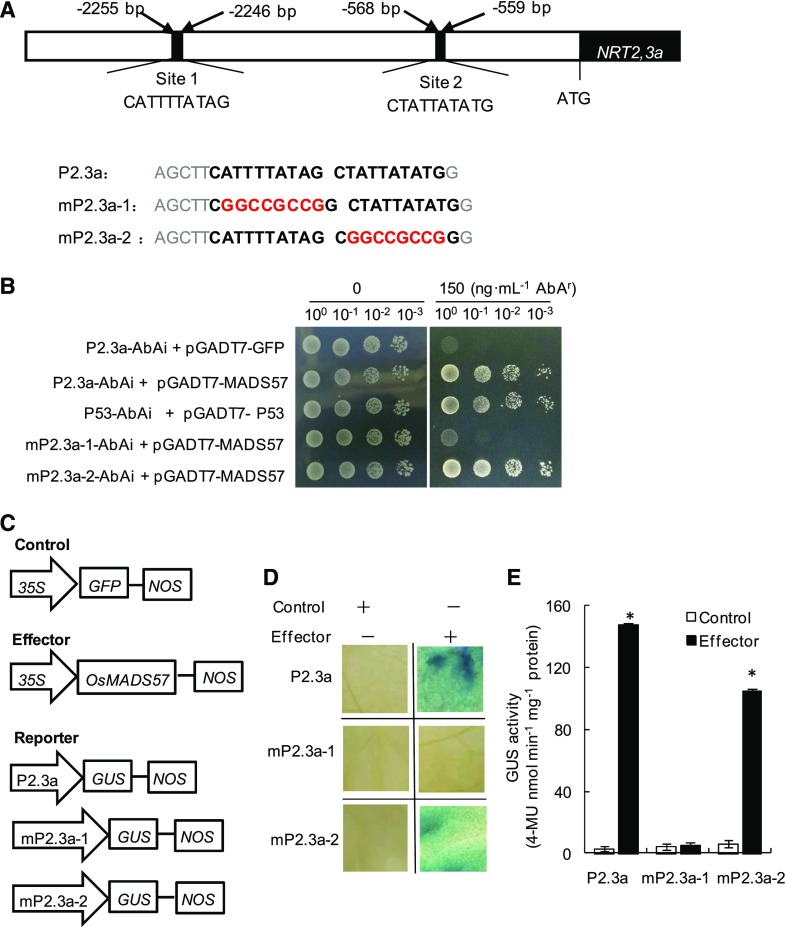
Figure 5.
OsMADS57 binds to the OsNRT2.3a promoter. A, Two CArG boxes (sites 1 and 2) in the OsNRT2.3a promoter. Letters in boldface indicate a tandem repeat of two potential CArG boxes in the promoter of OsNRT2.3a (P2.3a), and letters in red are the mutations of site 1 (mP2.3a-1) and site 2 (mP2.3a-2). B, Yeast cells were cotransformed with a bait vector containing a promoter fragment in A fused to an AbAi reporter gene and a prey vector containing MADS57 fused to a GAL4 activation domain. Cells were grown in liquid medium to an OD600 of 1 diluted to 0.1 and diluted in a 10× dilution series (from 10−1 to 10−3). From each dilution, 6 μL was spotted on medium selecting for both plasmids and selecting for interaction (synthetic dropout, –Ura, –Leu), supplemented with 150 nm AbA to suppress background growth and to test the strength of the interaction. C, Schematic diagram of the effector and reporter used for transactivation studies. The plasmid 35S::OsMADS57 was used as the effector, the plasmid P2.3a::GUS and its mutant versions mP2.3a-1::GUS and mP2.3a-2::GUS were used as the reporter, and 35S::GFP was used as an internal control. D and E, Transactivation activity was detected by GUS staining (D) and quantitative analysis of the GUS activity (E) after reporter and effector plasmids were coinfiltrated into N. benthamiana. Data in E are means of five replications ± se. *, P < 0.05 (Student’s t test) compared with the control. 4-MU, 4-Methylumbelliferone.