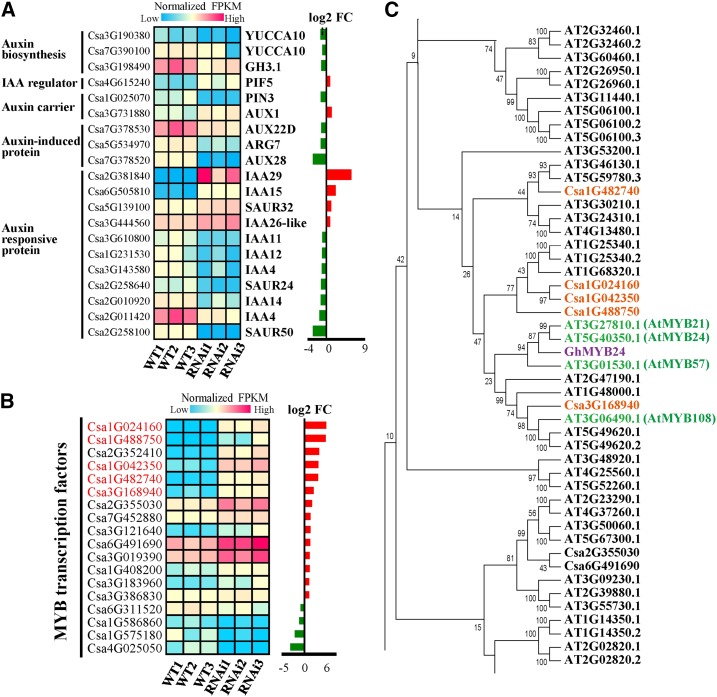
Figure 6.
Identification of auxin signaling and MYB transcription factor genes whose expression patterns were changed in flowers of CsSUT1-RNAi compared to wild-type (WT) plants. A, Heat map of DEGs involved in auxin metabolic pathways, including auxin biosynthesis, transport, and response. B, Heat map showing normalized FRKM values of MYB transcription factors; five genes that changed significantly are highlighted in red. C, Phylogenetic analysis of MYB transcription factors. The five cucumber proteins highlighted in red font shared high sequence similarity with Arabidopsis AtMYB21, 24, 57, 108 (green font), and cotton GhMYB24 (purple font). The genes presented in (A) and (B) were selected with an FPKM value ≥ 10 before down-regulation or after up-regulation. Abbreviations: FC, fold change; PIN3, PIN-formed 3; PIF5, phytochrome-interacting factor 5; AUX1, auxin transporter-like protein; FPKM, fragments per kilobase of transcript per million fragments mapped.