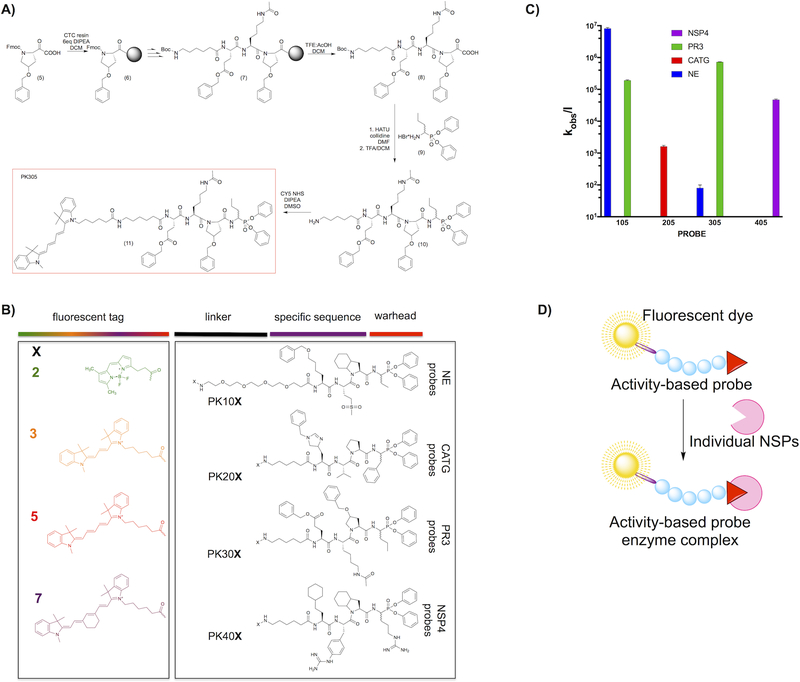
Figure 2. (a) Scheme of activity-based probe synthesis using PK305 as an example.
The respective P1 amino acid was loaded on the Chlorotrityl resin using DIPEA in dry DCM, followed by coupling of the P2–P4 residues by standard solid phase elongation. Following deprotection each tetrapeptide was cleaved from the resin with and attached to the hydrobromide salt of NvaP(OPh)2. The Boc protecting group was removed, followed by lyophilization and attachment of Cyanine5-NHS. The crude product was purified by HPLC, lyophilized and stored at −80°C until use. (b) Structures of the components of each activity-based probe used in this study. We utilized 16 fluorescent probes, and they are coded throughout the manuscript according to the N-terminal fluorescent tag (X) and specificity sequence as indicated in the panel. Thus, NE series probes are always PK10X, where X is one of the four fluorophores in the panel, CATG are PK20X, etc. for the other enzyme probes. (c) Second order rate constants for each Cy5 coded champion probe (PKX05 series) with each NSP. The log scale compresses the S.D. error bars from biological replicates, which were all within 10% of the mean. (d) The overall principle of activity-based probes.