Figure 7. A global model for how nuclear bodies shape overall three-dimensional genome organization in the nucleus.
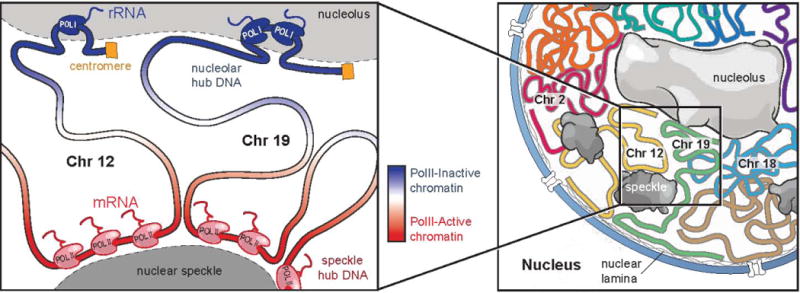
Left Panel: DNA regions containing a high-density of PolII associate with the nuclear speckle, while genomic regions linearly close to ribosomal DNA or centromeric regions associate with the nucleolus. This leads to co-association of multiple DNA regions around the same nuclear body to create spatial hubs of inter-chromosomal contacts. In addition to the genomic regions directly associating around these nuclear bodies, other DNA regions exhibit preferential organization, such that regions with higher levels of PolII density are closer to the nuclear speckle (red gradient) and regions with lower levels of PolII density are closer to the nucleolus (blue gradient). Right panel: These overall constraints act to shape the global layout of genomic DNA in the nucleus. DNA regions on the same chromosome tend to be closer to each other (colored lines). Yet, regions on different chromosomes containing similar properties organize around a nuclear body and can be closer to each other than to other regions contained on the same chromosome.
