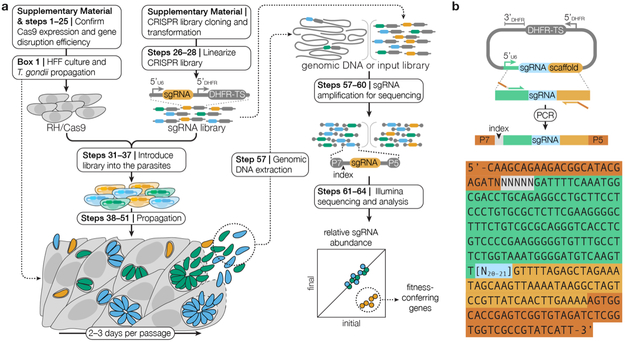
Figure 1. A Schematic of the CRISPR Screening Procedure.
(a) RH/Cas9 parasites that have been shown to express active Cas9 are transfected with the linearized CRISPR library. After one week of growth in HFF cells, transfected parasites may be analyzed to identify fitness-conferring genes or subjected to additional selective pressures prior to analysis. Genomic DNA is extracted from the post-growth and post-selection populations and sgRNAs are amplified from this genomic DNA and from the input library through two sequential PCRs. The Illumina sequencing adaptors P5 and P7, as well as indices that facilitate multiplexing, are added during the second PCR. The abundances of sgRNAs in the transfected library relative to all subsequent samples are quantified using next-generation sequencing. Genes that confer fitness will be lower abundance in post-growth or post-selection populations relative to the transfected CRISPR library. (b) Diagram of the amplification of sgRNAs from the CRISPR library prior to sequencing. The sequence of the amplicon is illustrated with N’s denoting the barcode for multiplexing (gray) and the sgRNA (blue). Regions or homology to the vector (orange and green), and Illumina sequencing adaptors (red) are also highlighted.