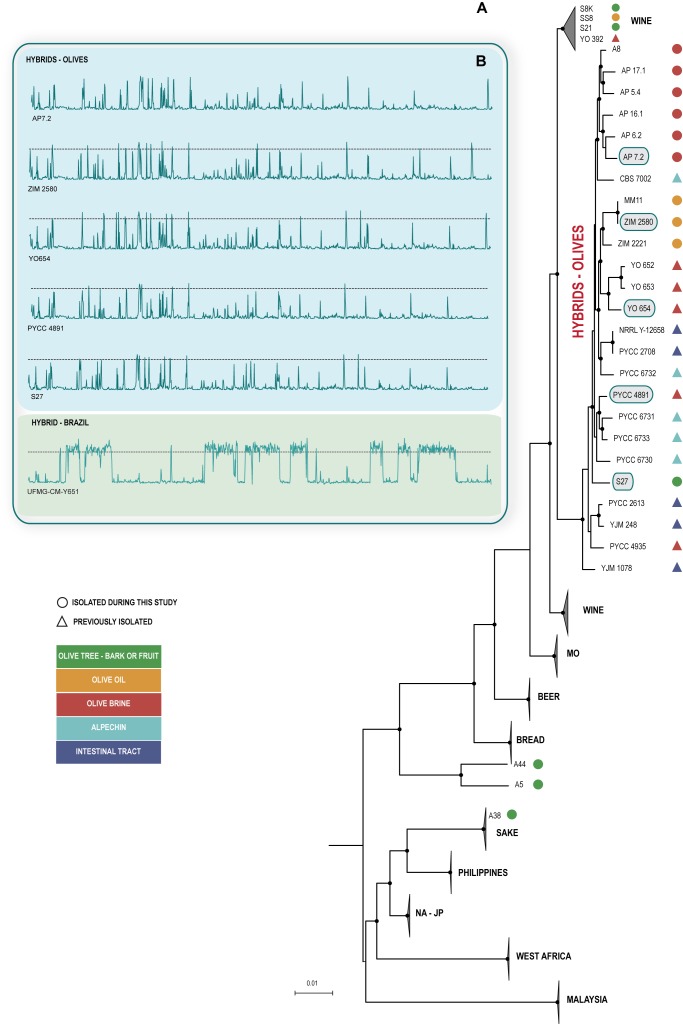
FIGURE 1.
Hybrid olive strains form a monophyletic group and have a similar genomic organization. (A) Phylogenetic placement of hybrid olive strains among the known lineages of Saccharomyces cerevisiae (MO, Mediterranean oaks; NA-JP, North America – Japan). Whole-genome phylogenetic tree constructed after discarding S. paradoxus regions in all genomes. The phylogeny was inferred from of 93 sequences and 913590 SNPs using the Maximum Likelihood method as implemented in IQ-TREE with the TVM+F+G4 model of sequence evolution and was rooted with S. paradoxus. Branch lengths correspond to the expected number of substitutions per site and black dots in tree nodes depict bootstrap support values above 85% (1000 replicates). Strains isolated from the olive niche are distinguished based on the specific isolation source (see color codes). (B) Similar divergence plots of the genomes of selected hybrid strains (highlighted in the phylogeny) to the reference genome of S. paradoxus CBS 432. The dotted lines depict the 10% divergence threshold that represents the average divergence between S. cerevisiae and S. paradoxus. The substantially distinct divergence plot of a Brazilian S. cerevisiae × S. paradoxus hybrid strain (UFMG-CM-Y651) previously reported by us (Barbosa et al., 2016) is included for comparison.