Figure 2.
Verification of Versatile Base Editing of ABEmax-NG Using EGFP Reporter System
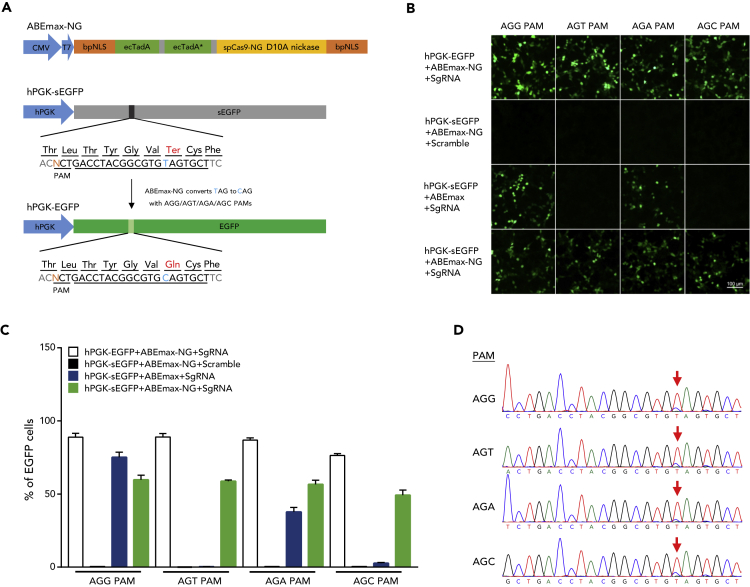
(A) ABEmax-NG construct and the schematic of the EGFP reporter system. The TAG stop codon was converted to glutamine by ABEmax-NG to restore EGFP protein production. The different third base in PAM does not change the original threonine of EGFP protein.
(B) ABEmax-NG-induced conversion of sEGFP to EGFP in HEK293FT cells. Scramble, non-targeting sgRNA served as negative control; sgRNA, sgRNA targeting sEGFP. Scale bars, 100 μm.
(C) Analysis of base editing by fluorescence-activated cell sorting. The percentage was calculated as the number of EGFP-positive cells/total transfected cells. Data are represented as the mean ± SEM (n = 3 from three independent experiments).
(D) The chromatograms of Sanger sequencing showing ABEmax-NG can induce sEGFP to EGFP conversion in different NG(N) PAM.