Figure 4.
ABEmax-NG Mediates Pathogenic Exon Skipping In Vivo
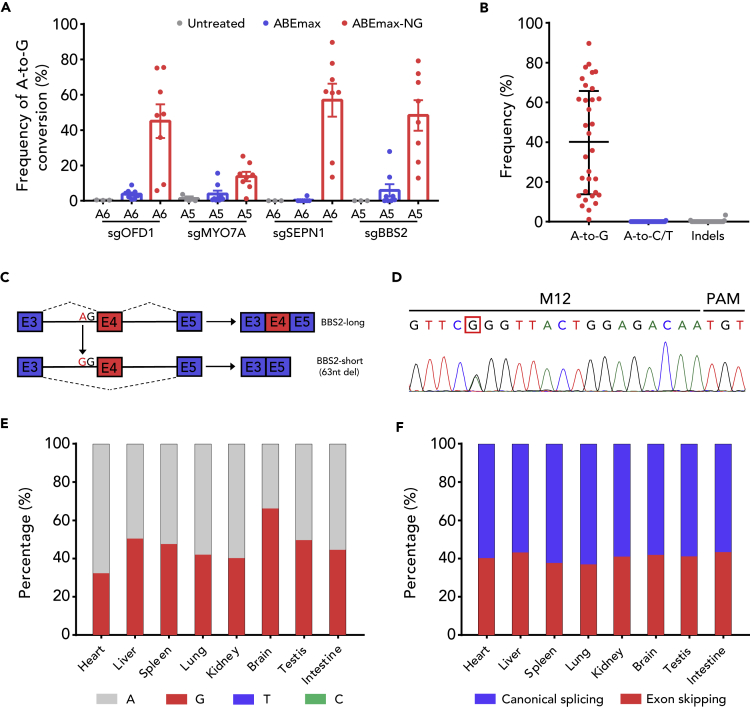
(A) A-to-G editing efficiency of ABEmax and ABEmax-NG at four splice sites in mouse embryos. Data are represented as the mean ± SEM (n = 3 for untreated, n = 8 for ABEmax, n = 8 for ABEmax-NG).
(B) Statistical analysis of the A-to-G editing frequency, unwanted base conversions, and indels induced by ABEmax-NG in (A). The median and interquartile range are shown.
(C) Schematic of ABEmax-NG-induced BBS2-long to BBS2-short switch. Top panel: Exon 4 is included in normal BBS2. Bottom panel: ABEmax-NG mutates the invariant A to G at the splice acceptor site, leading to the exclusion of Exon 4.
(D) The chromatogram of Sanger sequencing of tail confirmed heterozygous genotype of founder mouse M12.
(E) The individual fraction of each base induced by ABEmax-NG in BBS2 target splice site from different tissues of founder mouse M12 at 4 weeks. Data are analyzed by deep sequencing.
(F) Quantification of the rate of exon skipping of BBS2 Exon 4 from different tissues of founder mouse M12 at 4 weeks by deep sequencing of RT-PCR amplification of the cDNA.