Figure 3.
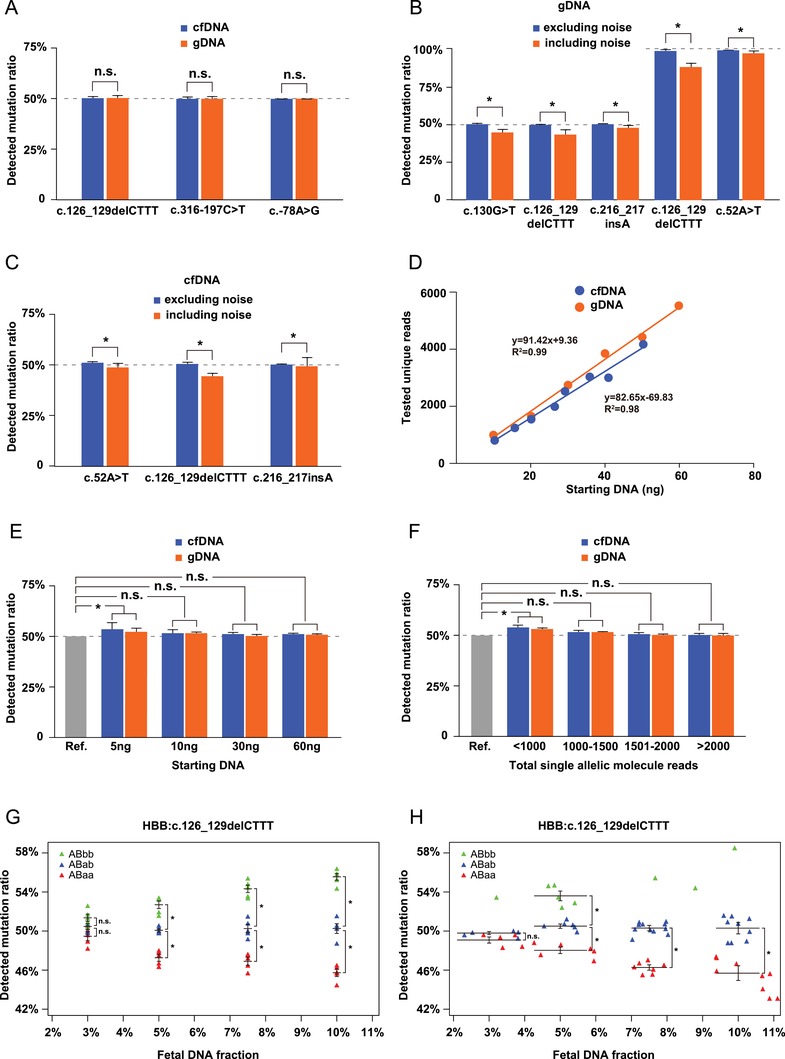
Development and optimization of the cfBEST system using β‐thalassemia as a model. A) Evaluation of gDNA as a reference sample. There was no significant difference between cfDNA and gDNA tested as a reference material with three different types of heterozygous β‐thalassemia mutations. Data are means ± SD; n = 5; n.s., not significant (Student t‐test). Evaluation of the impact of eliminating noise sequences through primer design and bioinformatic analysis with B) gDNA and C) cfDNA samples. A comparison of the detected mutation ratios between including and excluding noise sequences using B) gDNA samples from three heterozygous and two homozygous β‐thalassemia mutations and C) cfDNA samples from three heterozygous β‐thalassemia mutations. Data are means ± SD; n = 5; * p < 0.05, n.s., not significant (Student t‐test). D) Evaluating the correlation between starting DNA and tested unique reads (Pearson correlation coefficient analysis). E) Determining the minimal amount of starting DNA required for the cfBEST assay. Different amounts of gDNA and cfDNA from a heterozygous carrier of HBB:c.79G>A was tested as starting DNA. Data are means ± SD; * p < 0.05; n = 5; n.s., not significant (Student t‐test). All comparison was done between the theoretical value 50% (gray bar, denoted as a reference indicator, Ref.) and the detected ratios. F) Determining the minimal single‐molecule sequencing reads required for the cfBEST assay. The gDNA and cfDNA of a heterozygous carrier for HBB:c.79G>A was tested using cfBEST and different depths of sequencing reads were analyzed. Data are means ± SD; * p < 0.05; n = 5; n.s., not significant (Student t‐test). All comparison was done between the theoretical value 50% (gray bar, denoted as a reference indicator, Ref.) and the detected ratios. G) Determining the minimal fetal DNA fraction in maternal plasma required for accurate quantitative genotyping of β‐thalassemia mutations using ultrasonically fragmented gDNA by preparing the mixtures of different ratios. A mixture was made up of the sonicated gDNA from a heterozygous mutation sample that mimicked the background maternal cfDNA (denoted as “AB”) and “fetal” DNA sample (denoted by “aa” for a wild‐type, “ab” for a heterozygote, or “bb” for a homozygote). Four different concentrations of five replicate gDNA samples of ABaa, ABab, and ABbb were applied to cfBEST for mutation ratio detection. Data are means ± SD; n = 5. H) A total of 67 samples with HBB:c.126_129delCTTT, including 27 cases of ABaa, 31 cases of ABab, and 9 cases of ABbb from the peripheral blood of pregnant women were used to determine the lower limit of fetal DNA fraction. Different concentrations of cfDNA samples of ABaa, ABab, and ABbb were applied to cfBEST for mutation ratio detection. As there were no sufficient ABbb samples for statistics, individual dots denoted the detected ratios. Green triangles denote ABbb, blue triangles denote ABab, and orange triangles denote ABaa in (G,H).
