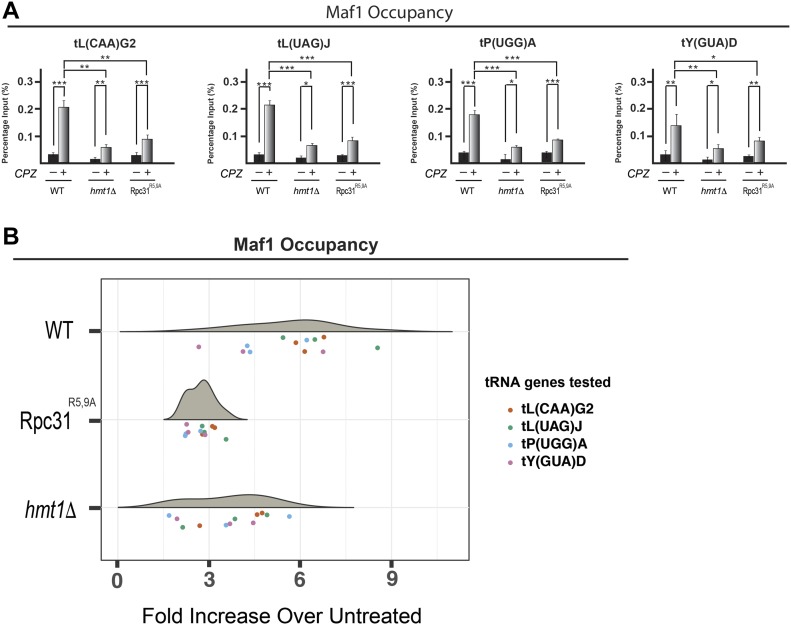
Figure 6. Decreased Maf1 occupancy at tRNA genes is observed in hmt1Δ and Rpc31R5,9A cells under stress.
(A) Maf1 occupancy across the four tRNA genes was determined by ChIP. qPCR results for products of ChIP performed on WT, hmt1Δ, or Rpc31R5,9A cells before and after treatment of CPZ are displayed as bar graphs. Percentage of input is calculated by ΔCT. The error bars representing SEM of three biological samples (n = 3). P-value as calculated by t test: *<0.05; **<0.01, and ***<0.001. (B) Fold increase in Maf1 occupancy of four candidate tRNA genes in WT, hmt1Δ, or Rpc31R5,9A cells after treatment with CPZ. qPCR was performed on products of ChIP in WT, hmt1Δ, or Rpc31R5,9A cells before and after treatment with CPZ. Percentage of input was calculated as ΔCT. ANOVA on these values revealed significant variation among the three strains (P-value = 4.6 × 10−6). Post hoc Tukey test revealed significant difference between WT and hmt1Δ (adjusted P-value is 8.6 × 10−4), and between WT and Rpc31R5,9A (adjusted P-value is 3.7 × 10−6). n = 3 per tRNA gene.