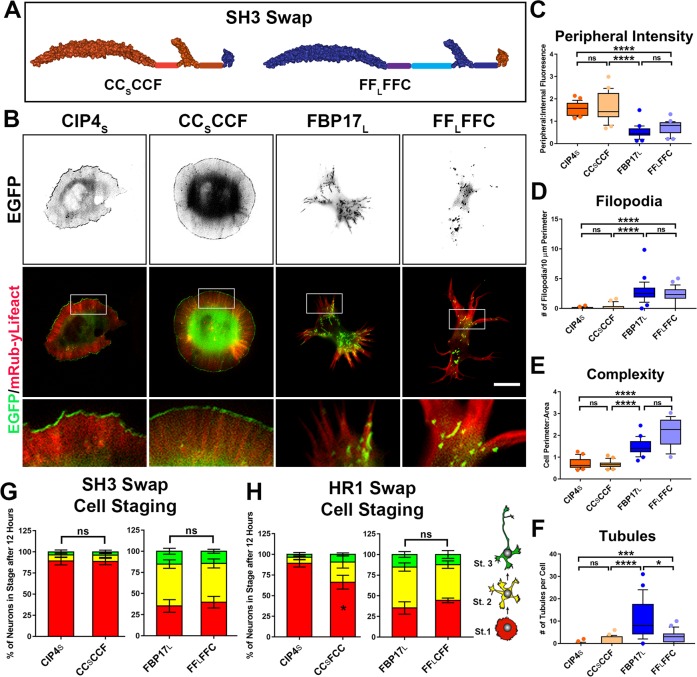
Figure S2. The SH3 domain has little effect on localization and function of CIP4 and FBP17.
(A) Schematic representation of the SH3 domain swap chimeras CCSCCF and FFLFFC. (B) Images of cortical neurons cotransfected with mRuby-Lifeact and either CIP4S-EGFP, CCSCCF-EGFP, FBP17L-EGFP, or FFLFFC-EGFP. (C–F) Box-and-whisker plots showing quantification of stage 1 neurons comparing the effects of the SH3 domain swap constructs on peripheral intensity (C), filopodia number (D), cell complexity (E), and tubule number (F). CIP4S-EGFP (n = 24 cells), CCSCCF-EGFP (n = 23 cells), FBP17L-EGFP (n = 24 cells), or FFLFFC-EGFP (n = 26 cells). One-way ANOVA with Kruskal–Wallis post-test multiple comparisons. (G) Stacked bar graph comparing the percentage of neurons in stage 1, 2, and 3 for neurons expressing CIP4S-EGFP (n = 50) versus CCSCCF-EGFP (n = 49) and FBP17L-EGFP (n = 61) versus FFLFFC-EGFP (n = 51) at 12 h postplating. (H) Stacked bar graph comparing the percentage of neurons in stage (st) 1, 2, and 3 in the HR1 domain swap constructs for neurons expressing CIP4S-EGFP (n = 50), CCSFCC-EGFP (n = 53), FBP17L-EGFP (n = 61), and FFLCFF-EGFP (n = 57) at 12 h postplating. Two-way ANOVA with Bonferroni post-test multiple comparison. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001; ns, not significant. Scale bars represent 5 µm in whole-cell images and 1 µm in insets.