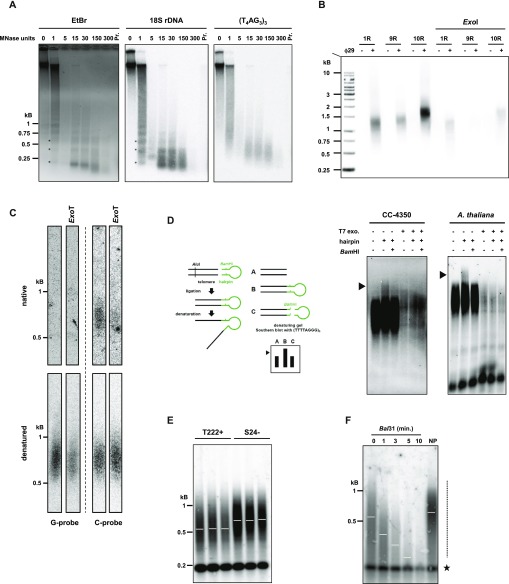
Figure 1. Structural characterization of C. reinhardtii telomeres.
(A) Characterization of C. reinhardtii telosomes by MNase digestion of chromatin (left panel; “EtBr”: ethidium bromide staining of the migration gel) and Southern analysis with a telomeric specific probe (right panel; (TTTTAGGG)3: radiolabeled probe). The membrane was then stripped and rehybridized with an 18S rDNA probe (middle panel). Lanes correspond to increasing amounts of MNase units; “Pr”: control where nuclei were digested with 60 MNase units after protein removal. (B) PETRA was used to amplify three different telomeres (1R, 9R, and 10R) from strain CC4350+ and analyzed by Southern blot hybridization using the telomere-specific probe (TTTTAGGG)3(see also Fig S1C). Negative controls include omission of the primer extension step by Φ29 polymerase and pretreatment of the samples with ExoI. (C) Upper panel: Native in-gel hybridization assay of telomeres from strain T222+ using a G-probe (oT0958, left) or a C-probe (oT0959, right). Most of the native signal, when hybridized with the C-probe, was absent when the gDNA was pretreated with ExoT. Lower panel: The native gel was then denatured and transferred to a membrane, which was then hybridized with the same probes. The uncropped gel and membrane are shown in Fig S1D. (D) Hairpin ligation assay on C. reinhardtii CC4350+ strain and A. thaliana. A scheme of the assay is shown (left). Digestion with BamHI, which removes the ligated hairpin and pretreatment with T7 exonuclease (“T7 exo.”), which resects the 5′ end of a duplex DNA, are used as controls. (E) T222+ and S24− strains were subcloned and three subclones were independently grown in liquid culture until stationary phase and subsequently analyzed by TRF Southern blot hybridization. (F) gDNA was subjected to Bal31 digestion for 0 to 10 min. Digested products were column-purified and then processed for TRF analysis. 0: no Bal31 digestion, but gDNA was column-purified before digestion by the restriction enzymes. NP: gDNA was directly analyzed by TRF, with No column Purification. Dashed line: smear corresponding to telomeres. Star: Bal31-insensitive band, corresponding to interstitial telomeric repeat (see also Fig S1E).