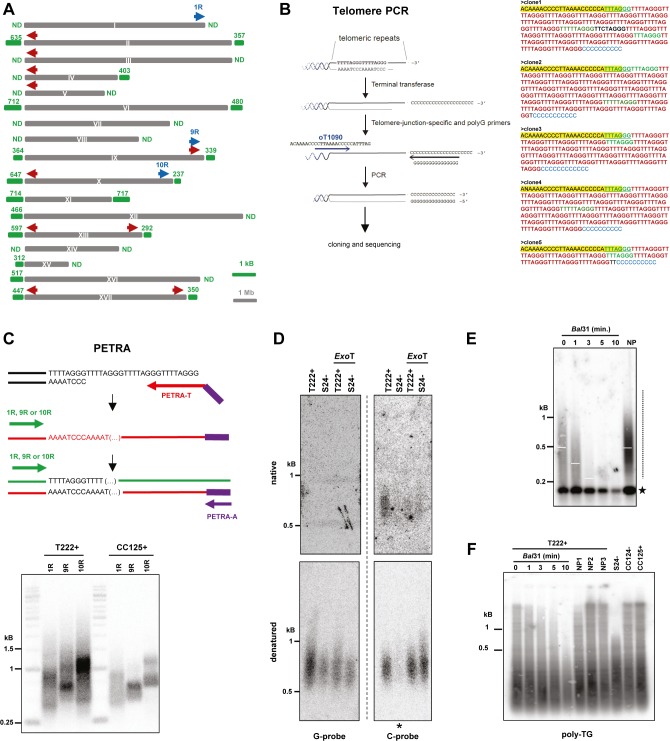
Figure S1. Characterization of C. reinhardtii telomere sequence, end structure, and length distribution. Related to Fig 1.
(A) Scheme of the 17 C. reinhardtii chromosomes. Red arrow: forward primer oT1090 used in telomere PCR. Blue arrows: forward primers 1R, 9R, and 10R used in PETRA experiments. Green numbers refer to telomere length from the public genome sequence of C. reinhardtii (Phytozome genome version 5.5; https://phytozome.jgi.doe.gov/pz/#!info?alias=Org_Creinhardtii). ND: no telomere in the database. (B) Scheme of the different steps of telomere PCR (left). After adding a poly-C extension to telomeric ends by terminal transferase, PCR amplification using a forward primer at a subtelomere-telomere junction (oT1090) and a poly-G reverse primer was performed. Telomere PCR was used to amplify telomeric repeats from the T222+ strain. The resulting PCR products were cloned in E. coli and 32 clones sequenced. Five examples of telomere PCR sequences showing the sequence of primer oT1090 (yellow), the poly-C extension (blue), canonical C. reinhardtii repeats (TTTTAGGG, in red), and variant motifs (other colors) (right). The frequency of the observed repeats is indicated in Table 1. (C) Top: Scheme of the PETRA assay for 3′ overhang detection. The primer PETRA-T was annealed to the putative 3′ overhang and extended using DNA Φ29 DNA polymerase. PCR amplification was then performed using a subtelomere-specific primer and PETRA-A primer, which annealed to the nontelomeric sequence of the 5′ end of PETRA-T. Bottom: PETRA performed on strains T222+ and CC125+ for three telomeres (1R, 9R, and 10R). (D) Uncropped images corresponding to the in-gel hybridization assay shown in Fig 1C. Native gels (upper panel) and denatured signal detected on membrane (lower panel) are shown for both the G-probe and the C-probe. Strain S24− was cropped out for Fig 1C because of an underloaded lane (asterisk). (E) Independent biological replicate of the Bal31 assay performed on strain T222+ (see Fig 1F). (F) A poly-TG probe did not detect the band observed at ∼200 bp with a telomere probe. gDNAs from strains T222+, S24−, CC124− and CC125+ were processed as for TRF experiments, Southern blotted, and then hybridized with a radioactive (TG)13 probe. T222+ samples were also treated by Bal31 exonuclease for increasing amounts of time (five first lanes) before digestion with the restriction enzymes. NP1-3 samples were not column-purified.