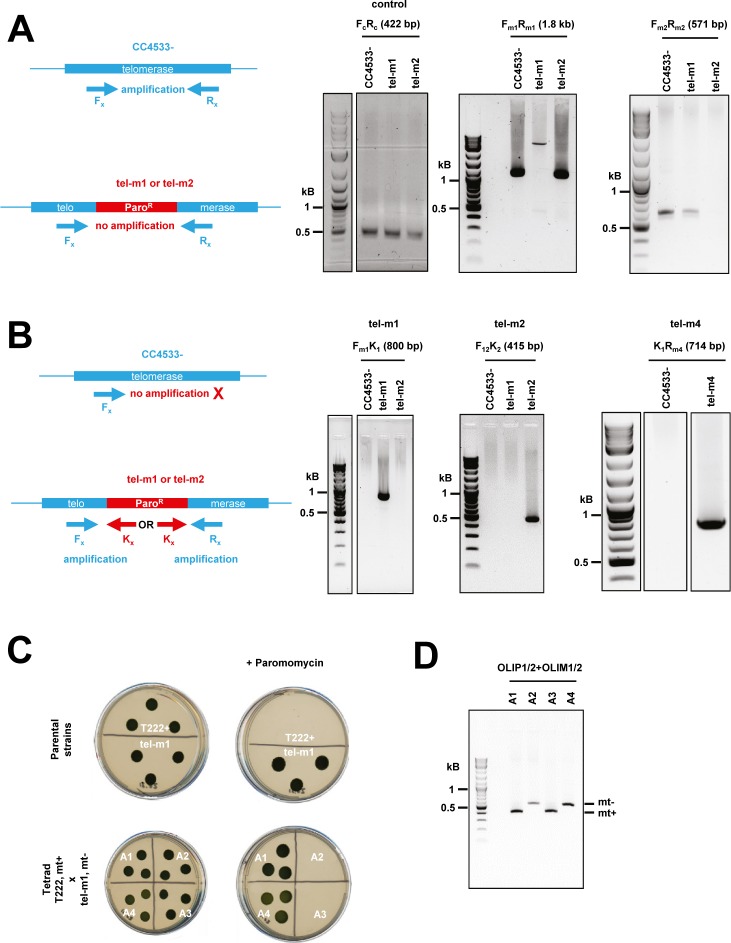
Figure S4. PCR verification of insertion mutants tel-m1 and tel-m2. Related to Fig 4.
(A) Scheme of expected PCR amplification product (left). Fx and Rx denote primers where “x” stands for “m1” or “m2.” PCR using primers specific to the reported insertion sites in tel-m1 (Fm1 and Rm1) and tel-m2 (Fm2 and Rm2) gave no signal in the targeted mutant because the insert is too large for efficient amplification but amplified the expected product in the reference strain and the other mutant (of size 1.8 kb and 571 bp, respectively; gels in the middle and on the right). Primers Fc and Rc amplified a 422-bp product in the CrTERT gene outside of the predicted insertion sites in tel-m1 and tel-m2 (positive control, gel on the left). (B) Scheme of expected PCR amplification product using specific primers for tel-m1 (Fm1) and tel-m2 (F12), combined with primers in the paromomycin cassette (K1 and K2) (left). The two pairs of primers amplified the expected products in the respective mutants (right). All bands were excised, gel-purified, and sequenced. PCR amplification using primers K1 and Rm4 were used to characterize the insertion in tel-m4. The specific band obtained in tel-m4 was excised from the gel and sequenced and could indeed be mapped to the expected CrTERT sequence. (C) Mutant tel-m1 was crossed with T222+ strain. The cross with tel-m2 is not shown. The parental strains (tel-m1 and T222+) as well as the progeny of the four haploids of a tetrad from the diploid are spotted on TAP solid media with (“+ Paromomycin”) or without paromomycin (10 μg·ml−1). Five tetrads were analyzed for each cross for the segregation of paromomycin resistance. Mating types “mt+” and “mt−” are indicated. (D) The 2:2 segregation of the mating type (“mt+” and “mt−”) was checked by PCR.