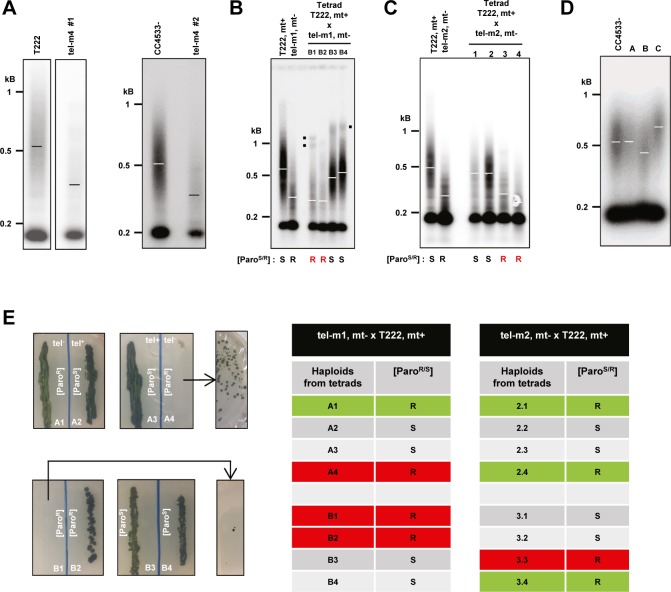
Figure S5. Telomere length and senescence phenotype of CrTERT mutants. Related to Fig 5.
(A) Insertion mutant tel-m4 has shorter telomeres than T222+ and CC4533− as shown by TRF analysis of two biological replicates of the tel-m4 mutant strain. (B) Independent tetrad of the tel-m1 x T222 cross analyzed by TRF Southern hybridization performed on the haploid progenies (see also Fig 5B). Mating types “mt+” and “mt−” are indicated. (C) Independent tetrad of the tel-m2 x T222 cross analyzed by TRF Southern hybridization performed on the haploid progenies (see also Fig 5C). Mating types “mt+” and “mt−” are indicated. (D) TRF Southern blots of three insertion mutants from the CliP library in genes not annotated to be telomere-related compared with the parent CC4533− strain. Mutant A = LMJ.RY0402.203280; mutant B = LMJ.RY0402.060361; mutant C = LMJ.RY0402.166033. (E) Telomerase-negative haploid progenies A1-4 and B1-4 are shown after about 6 mo of maintenance on solid media. A4 and B1 experienced growth defect and cell death typical of replicative senescence but generated clonal low-frequency survivors after additional restreaks. The telomerase-negative haploids A1 and B2 did not show senescence phenotype. Right: summary table of the presence (red) or absence (green) of senescence phenotype in telomerase-negative (paromomycin-resistant) progenies of tetrads from the indicated crosses.