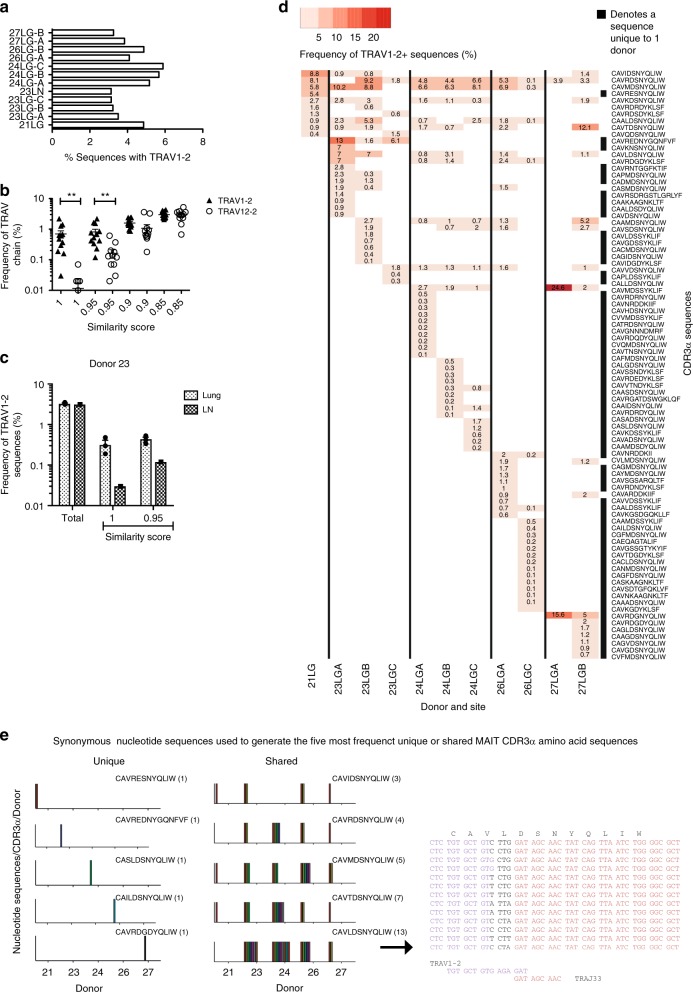
Fig. 2.
Expansions of MAIT cell-consistent CDR3α‘s are present in tuberculous lung granulomas. a Frequency of TRAV1-2+ sequences as a percentage of all productive TCRα sequences. In some cases, multiple areas of tissue were sampled, ranging from closest (A) to furthest (C) from the site of disease. b MAIT cell TCRα sequences are consistent with similarity scores of 0.95 and 1. Each symbol represents the frequency of TRAV1-2+ or TRAV12-2+ sequences within each similarity score for each donor sample (n = 12 biologically independent samples). c Frequency of total TRAV1-2+ sequences or those with similarity scores of 0.95 and 1 in the lung (n = 3 biologically independent samples) and mediastinal lymph node (LN, 1 sample) from donor 23. Height represents mean, error bars represent standard error. d Frequencies among TRAV1-2+ sequences of the top 10 public and private MAIT cell CDR3α sequences (MAIT Match score ≥0.95) across individual donors and lung samples. e Variation in the number of synonymous nucleotide sequences encoding the five most frequent private (left) and public (right) MAIT cell CDR3α amino acid (aa) sequences from all samples displayed in Fig. 3d. For each aa sequence, each colored bar represents a different nucleotide sequence. The 13 different nucleotide sequences used to generate the shared MAIT cell CDR3α aa sequence CAVLDSNYQLIW are displayed. Text color represents nucleotide origin: purple (TRAV), black (TRAD or n insertion), red (TRAJ). LG lung granuloma, LN lymph node. Source data are provided in Supplementary Data