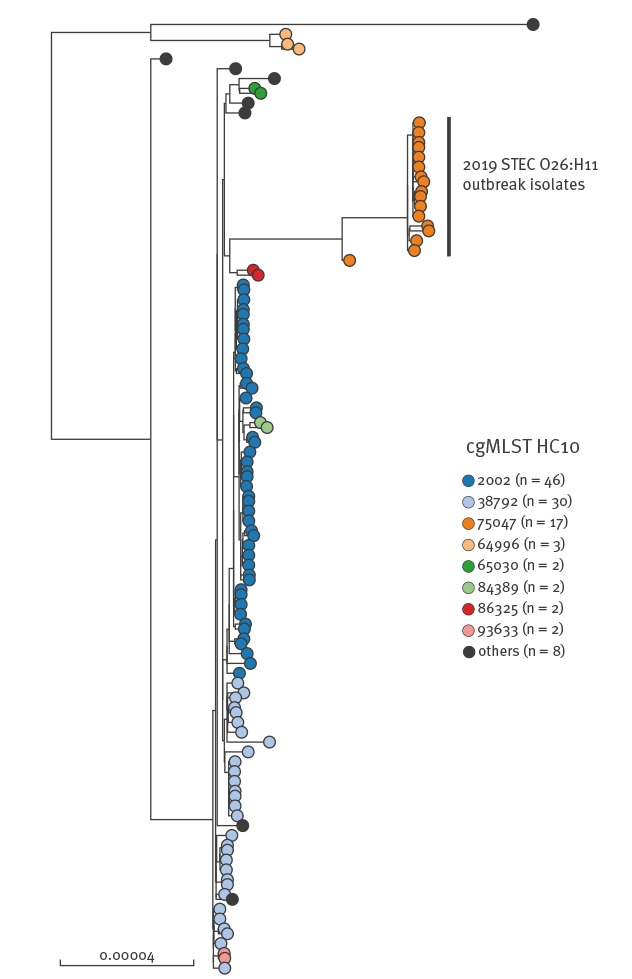
Figure 2.
SNP-based phylogenetic tree of STEC O26:H11, ST21, performed by the NRC-ESS, France, 2019
cgMLST: core genome multilocus sequence typing; NRC-ESS: French National Reference Centre for Escherichia coli, Shigella and Salmonella; STEC: Shiga-toxin producing Escherichia coli; SNP: single nucleotide polymorphism.
The phylogenetic analysis was carried out by using the « create SNP project » tool in Enterobase on 105 French STEC genomes belonging to cgMLST type HC20|2002, with the isolate 201902833 as reference. The tree was based on 2,463 SNPs found in non-repetitive regions that are present in 97% or more of all the queried genomes. The tips are coloured according to the cgMLST type HC10. The scale bar refers to the frequency of mutation per site in the core genome (this corresponds to approximately 180 SNPs).