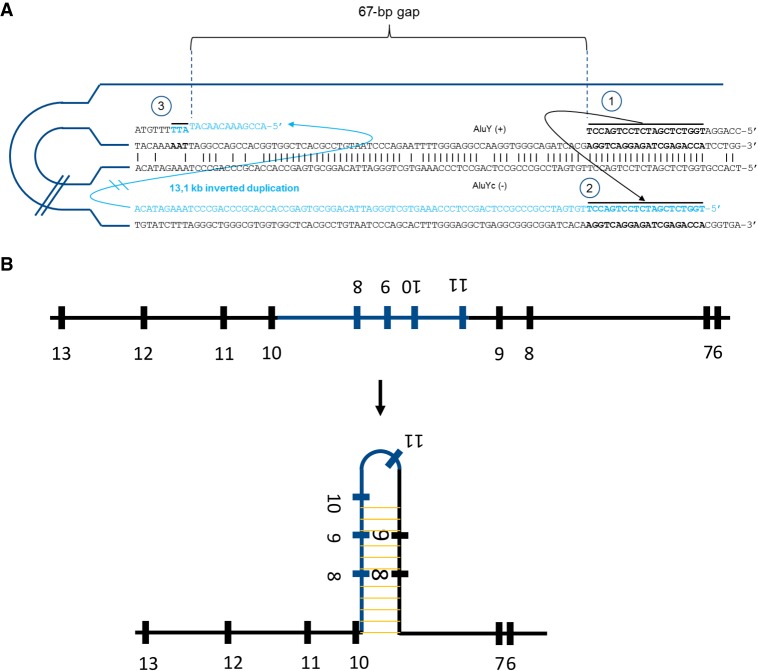
Figure 4.
Proposed mechanisms underlying the observed inverted duplication and exon skipping. (A) We propose the following sequence of events causing the inverted duplication: (1) replication stalls at AluY (+), possibly facilitated by the interaction with a second Alu element, AluYc, in opposite orientation; (2) template switching between both Alu elements takes place, which is mediated by a 20-base-long microhomology region and causes the large inverted duplication; and (3) replication switches back to the original template directly downstream from the highly homologous regions between the Alu elements, leaving a 67-base-long nonreplicated gap. This template switching is mediated by a 3-base-long area of microhomology. (B) The inverted duplication creates a large area of full complementarity within the pre-mRNA that induces the formation of a hairpin, which in turn forces the spliceosome to skip exons 8 and 9. Even proper splicing of exon 10 is compromised.