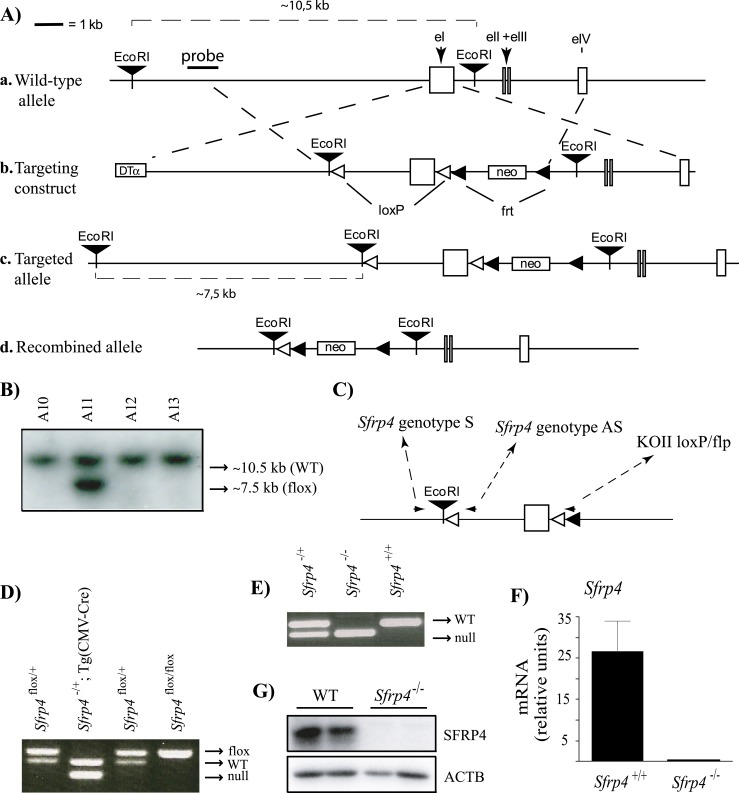
Figure 1.
Targeting of Sfrp4. (A) Illustration of the strategy used to generate Sfrp4−/− mice. (a) Placement of the genomic DNA probe used for Southern blotting. (b) In the targeting construct, LoxP sites were inserted upstream and downstream of the first exon of Sfrp4. (c) Sfrp4flox allele. (d) Sfrp4− (i.e., Cre-recombined) allele. (B) Generation of ES cell lines heterozygous for the targeted Sfrp4 allele. Presence of the targeted allele was detected as a 7.5-kb EcoRI restriction fragment by Southern blotting. One representative positive clone (A11) is shown alongside three negative clones. (C) PCR genotyping analysis strategy. Oligonucleotides (small black arrowheads) were designed to generate PCR products of 249 bp for the Sfrp4+ allele, 143 bp for the Sfrp4flox allele, and 136 bp for the Sfrp4− allele. (D) PCR genotype analysis of pups generated by the mating of Sfrp4flox/+ with Tg(CMV-cre)1Cgn/J mice. (E) PCR genotype analysis of pups generated by backcross of Sfrp4+/− mice. (F) Sfrp4 mRNA levels and (G) SFRP4 protein levels in GCs isolated from immature eCG-treated Sfrp4+/+ and Sfrp4−/− mice 12 h after the administration of an ovulatory dose of hCG, confirming the abrogation of SFRP4 expression in the Sfrp4-null mice. The mRNA levels were measured by RT-qPCR (n = 6 animals per genotype). Data are expressed as means ± SEM. DTα, diphtheria toxin α chain; neo, neomycin resistance cassette.