ABSTRACT
Signaling pathways that mediate cell-cell communication are essential for collective cell behaviors in multicellular systems. The hedgehog (HH) pathway, first discovered and elucidated in Drosophila, is one of these iconic signaling systems that plays many roles during embryogenesis and in adults; abnormal HH signaling can lead to birth defects and cancer. We review recent structural and biochemical studies that have advanced our understanding of the vertebrate HH pathway, focusing on the mechanisms by which the HH signal is received by patched on target cells, transduced across the cell membrane by smoothened, and transmitted to the nucleus by GLI proteins to influence gene-expression programs.
KEY WORDS: Cholesterol, Hedgehog signaling, Morphogen, Patched, Primary cilium, Smoothened
Summary: This Review discusses recent structural and biochemical studies that have advanced our understanding of the vertebrate hedgehog pathway, a cell-cell communication system that plays important roles in development and disease.
Introduction
Secreted hedgehog (HH) ligands are paracrine signaling factors that mediate communication between cells over distances as large as several hundred microns (Lewis et al., 2001). The first gene encoding a HH ligand was identified genetically through its role in patterning the Drosophila larval epidermis (Nusslein-Volhard and Wieschaus, 1980). Expansion of this gene family has produced three paralogs in amniotes: desert hedgehog (Dhh), Indian hedgehog (Ihh) and sonic hedgehog (Shh). Vertebrate HH ligands, like the Drosophila Hh protein, play roles in patterning multiple tissues including the limb bud, nervous system and skeleton (McMahon et al., 2003). HH ligands can drive proliferation or function as morphogens: secreted from organizing centers, they disperse to form spatial and temporal gradients that provide positional information across a field of progenitor cells to inscribe a pattern of cell fates on a developing tissue (Echelard et al., 1993; Krauss et al., 1993; Riddle et al., 1993; Roelink et al., 1994). HH signaling should be viewed as a system that drives distinct outcomes depending on the strength and duration of signaling activity in target cells, and not a binary ON/OFF switch. Indeed, even modest alterations in HH signaling strength can lead to human birth defects (Nieuwenhuis and Hui, 2005). The capacity for quantitative signaling might be an intrinsic consequence of the evolution of the HH pathway from an ancient system that sensed and regulated cellular metabolite levels (Bazan and de Sauvage, 2009; Hausmann et al., 2009). In addition to their roles during embryogenesis, HH ligands also function in paracrine signaling networks to regulate tissue homeostasis and regenerative responses in adults (Lee et al., 2016). Mutations in HH pathway components that increase signaling strength can drive tumorigenesis, and two HH pathway inhibitors are currently used in the clinic to treat basal cell carcinoma (Raleigh and Reiter, 2019).
The HH pathway (reviewed by Briscoe and Therond, 2013; Lee et al., 2016) is unusual among signaling systems in being composed of a series of inhibitory interactions (Fig. 1A). The main receptor for HH ligands is the 12-pass transmembrane (TM) protein patched (PTCH). In the absence of ligands, PTCH inhibits signaling by suppressing the activity of smoothened (SMO), a heptahelical TM protein that belongs to the G-protein coupled receptor (GPCR) superfamily (Ingham et al., 1991). When SMO is inactive, two inhibitory components – suppressor of fused (SUFU) and protein kinase A (PKA) – restrain the transcriptional activity of the GLI family of transcription factors by direct association and phosphorylation, respectively. Under the influence of SUFU and PKA, the GLI proteins undergo partial proteolysis into repressors (GLI-R) that enter the nucleus and suppress the transcription of target genes. HH ligands trigger serial dis-inhibition of steps in the pathway (Fig. 1A). They bind and inhibit PTCH, thus liberating SMO to adopt an active conformation. Activated SMO transmits the HH signal across the membrane and overcomes the negative influence of PKA and SUFU on GLI proteins. Instead of undergoing proteolytic processing, GLI proteins dissociate from SUFU, enter the nucleus and activate the transcription of target genes.
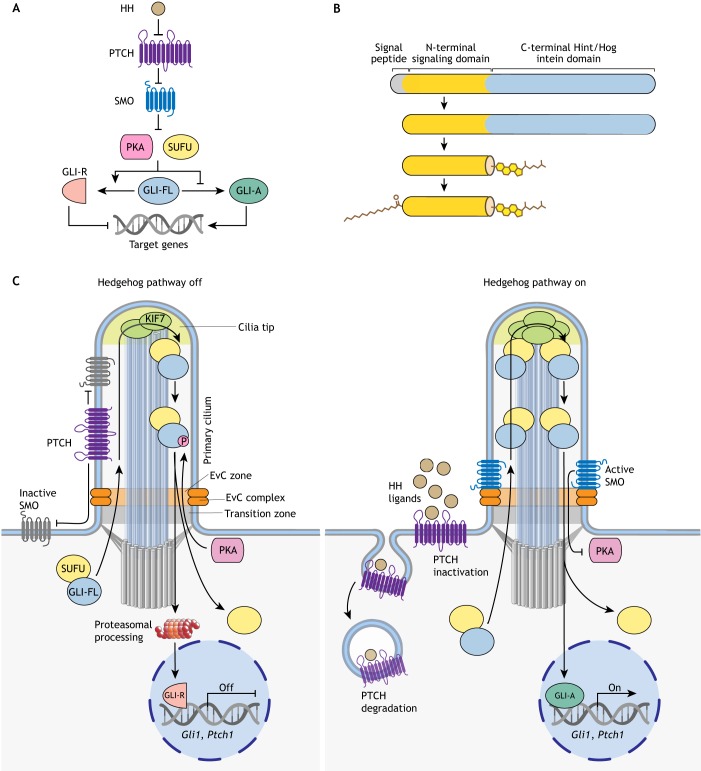
Fig. 1.
Overview of HH signaling. (A) HH signaling regulates a bi-functional transcription factor that can repress (GLI-R) or activate (GLI-A) the transcription of target genes. HH ligands bind and inhibit the function of their receptor PTCH, allowing SMO to adopt an active conformation. SMO transmits the HH signal across the membrane and antagonizes the function of two negative regulators, SUFU and PKA, which promote GLI-R formation. Consequently, full-length GLI proteins (GLI-FL) are converted to GLI-A. (B) All HH ligands are modified with a cholesteroyl group at their C termini, attached through an auto-proteolytic reaction catalyzed by the C-terminal domain, and a palmitoyl group at their N termini, attached by a membrane-bound O-acyltransferase. (C) Vertebrate HH signaling is associated with protein trafficking events at primary cilia. When the HH pathway is ‘off’ (left), PTCH is enriched in cilia and inhibits SMO. PKA and SUFU restrain GLI activity and promote its proteolysis into GLI-R. HH signaling is turned on in target cells (right) when HH ligands inhibit PTCH and induce its clearance from primary cilia. As a result, SMO is activated and accumulates in cilia in association with a scaffolding complex, the Ellis van Creveld (EvC) complex. Activated SMO antagonizes the inhibitory effect of PKA on the GLI proteins, leading to the dissociation of SUFU. Now, instead of being converted into GLI-R, GLI-FL can enter the nucleus and activate target gene transcription (GLI-A). The transition zone at the cilia base regulates receptor access to cilia, cilia tips form a compartment (marked by the kinesin KIF7) that regulates the GLI proteins, and the EvC complex scaffolds SMO signaling near the cilia base (Box 1).
HH signaling in vertebrates (but not in Drosophila) depends on primary cilia – solitary microtubule-based organelles that function as signaling hubs in development (Box 1) (Fig. 1C) (Huangfu et al., 2003). The connections between primary cilia and HH signaling are not our primary focus, and we refer the reader to a recent comprehensive review (Bangs and Anderson, 2017). Instead, we analyze recent progress in understanding the series of biochemical reactions that transmit the HH signal from the cell surface to the nucleus. We pay particular attention to recently reported structures of the TM proteins that detect HH signals at the cell surface and transmit them across the membrane to the cytoplasm. A theme that links our discussions is the regulation of signaling strength by HH morphogens. We do not discuss the myriad biological roles of HH signaling in development, cancer and regeneration, which is best left to a dedicated review. Finally, we focus on the vertebrate HH pathway, but note that HH signaling was discovered and elucidated by genetic and cell biological studies in Drosophila (recently recounted by Ingham, 2018).
Box 1. Primary cilia function as compartments for HH signaling.
In vertebrates, most HH pathway components are found localized within cilia, with transduction of the signal correlated with a set of choreographed protein trafficking events (Fig. 1C) (Corbit et al., 2005; Haycraft et al., 2005; Rohatgi et al., 2007). The seminal discovery that linked cilia to HH signaling came from mouse genetics, which identified a set of genes necessary for both cilia formation and signaling (Huangfu et al., 2003). More recently, genome-wide CRISPR-based screens in cultured cells confirm the inextricable link between primary cilia and HH signaling, identifying a multitude of cilia genes, the loss of which influences the strength of HH signaling in target cells (Breslow et al., 2018; Pusapati et al., 2018a). Although studies of primary cilia have transformed our view of vertebrate HH signaling, we still lack an understanding of which biochemical reactions occur in cilia and how these reactions are linked to cilia trafficking events. One emerging principle is that signaling reactions are compartmentalized in specialized microdomains within cilia: the transition zone (which regulates the entry of ciliary receptors), the cilia tip compartment (which regulates the activity of GLI proteins) and the EvC zone near the cilia base (which scaffolds SMO signaling) (Fig. 1C) (Dorn et al., 2012; Garcia-Gonzalo et al., 2011; He et al., 2014a; Pusapati et al., 2014). Such spatial segregation of signaling reactions, linked by transport mechanisms, might enhance the efficiency, specificity and directionality of signaling.
The biogenesis of HH ligands and their spread through tissues
HH ligands are synthesized as ∼45 kDa precursors that undergo an intein-like self-cleavage reaction, liberating an N-terminal signaling domain (HhN) covalently attached to a cholesteroyl moiety at the C terminus (Fig. 1B) (Lee et al., 1994; Porter et al., 1996). A palmitoyl moiety is then added to the N-terminus by the membrane-bound O-acyltransferase HHAT (Buglino and Resh, 2008; Chamoun et al., 2001; Pepinsky et al., 1998), generating the mature, dually lipidated ligand (Fig. 1B). The biogenesis, secretion and dispersal of HH ligands through tissues has been thoroughly reviewed recently (Manikowski et al., 2018; Petrov et al., 2017).
Regulation of secretion and transport is important for shaping temporal and spatial gradients of HH ligands across developing tissues, signaling strength in target cells and consequently tissue patterning outcomes. The dual lipidic modification on HH ligands render them highly hydrophobic and tethered to cell membranes. Short-range signaling between adjacent cells can be mediated by cell-surface-bound HH ligands (Strigini and Cohen, 1997). However, long-range signals require specialized components to release ligands from membranes and shield their lipidic appendages in the aqueous interstitial environment (Caspary et al., 2002; Lewis et al., 2001). The TM protein Dispatched (DISP1) is required exclusively for ligand release and likely functions to isolate HH ligands from the bulk membrane by binding to their cholesteroyl moieties and transferring them to a carrier for transport through tissues (Burke et al., 1999; Caspary et al., 2002; Kawakami et al., 2002; Ma et al., 2002; Tukachinsky et al., 2012). Several types of carriers have been identified, including the Scube family of secreted proteins, lipoproteins, extracellular vesicles and multimers of ligands themselves (summarized by Petrov et al., 2017). A distinct solution to the ligand transport problem is provided by cytonemes – long, actin-based cellular extensions that directly deliver ligands to distant cells without the requirement for ligand release from membranes (Bischoff et al., 2013; Chen et al., 2017; Ramírez-Weber and Kornberg, 1999; Sanders et al., 2013). Finally, there is a large body of work (summarized by Petrov et al., 2017) showing that heparan sulfate proteoglycans (HSPGs) can regulate the release, dispersal and reception of HH ligands.
PTCH is the main receptor for HH ligands
All HH ligands must bind and inhibit the function of PTCH to trigger signaling in target cells (Marigo et al., 1996a; Stone et al., 1996). Vertebrates have two PTCH genes, PTCH1 and PTCH2, but PTCH1 is the major regulator of signaling in vivo (Carpenter et al., 1998; Motoyama et al., 1998; Nieuwenhuis et al., 2006). We use PTCH to refer to both for simplicity. PTCH plays two separate roles in HH signaling: it inhibits SMO and reduces the abundance of HH ligands by promoting their endocytosis and lysosomal degradation (Chen and Struhl, 1996). PTCH gene expression is directly activated by HH signaling, resulting in a negative feedback loop: HH ligands inactivate PTCH and de-repress SMO, causing increased production of PTCH, which then feeds back both to inhibit SMO and reduce the abundance of HH ligands (Goodrich et al., 1996; Marigo et al., 1996b). A consequence of this negative feedback is that different doses of HH ligands are translated into different durations of signaling in target cells, which activates different sets of target genes (Dessaud et al., 2007). A combination of in vitro morphogen modeling in cultured fibroblasts and computational simulations show that several unique properties of PTCH are important for generating stable signaling gradients (Li et al., 2018). Such properties include negative feedback, inhibition of HH ligands and SMO, and involvement in a ‘double-negative’ circuit (in which SHH inhibits PTCH, which inhibits SMO).
PTCH has homology with two membrane transporter types: the resistance-nodulation-division (RND)-family pumps, which use transmembrane proton gradients to efflux toxic molecules out of gram-negative bacteria, and Niemann-Pick C1 (NPC1), which transports cholesterol from the lumen of the lysosome to the cytoplasm (Carstea et al., 1997; Loftus et al., 1997; Pfeffer, 2019; Tseng et al., 1999). Recently published cryo-electron microscopy (cryo-EM) structures reveal that PTCH resembles RND transporters and is composed of two pseudo-symmetrical segments (Fig. 2A) (Gong et al., 2018; Qi et al., 2018a,b; Zhang et al., 2018). Each segment includes a transmembrane domain (TMD) composed of six TM helices and one extracellular domain (ECD) interposed between TM1 and TM2. Likewise, NPC1 is also related to RND transporters, but notably transports cholesterol in the opposite direction to bacterial RND proteins. In PTCH, TM2-TM6 forms a sterol sensing domain (SSD; dark blue in Fig. 2A), which is also found in NPC1 and other proteins that handle cholesterol (Davies and Ioannou, 2000). The similarities between PTCH, NPC1 and RND proteins suggest that PTCH transports a hydrophobic ligand for SMO.
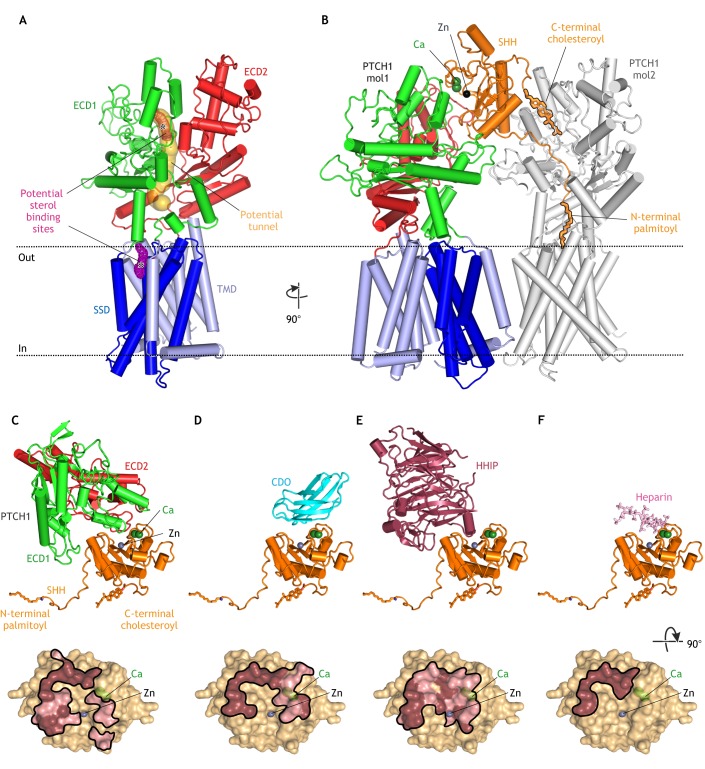
Fig. 2.
Structures of PTCH. (A) Structure of unliganded PTCH (PDB 6DMB; Gong et al., 2018) showing the transmembrane domain (TMD), which includes a sterol-sensing domain (SSD), and two extracellular domains (ECD1 and ECD2). A possible hydrophobic tunnel (ocher surface) is shown connecting two putative sterol-binding sites (meshed surfaces, asterisks) in ECD1 and the SSD. (B) Structure of the asymmetric 1SHH:2PTCH complex (adapted from PDB 6E1H; Qi et al., 2018b) reveals two distinct SHH-PTCH interfaces. PTCH1 molecule 1 (mol1) binds to SHH at an interface including its calcium- and zinc-binding sites and PTCH1 molecule 2 (mol2) engages the N-terminal palmitoyl and C-terminal cholesteroyl modifications of SHH, which are inserted into the PTCH protein core. The interaction of SHH with mol1 drives PTCH endocytosis and the palmitate-centered interaction with mol2 inactivates the transporter function of PTCH. (C-F) Structures of SHH in complex with the PTCH ECD (C; adapted from PBD 6E1H), CDO (D; PDB 3D1M; McLellan et al., 2008), HHIP (E; PDB 2WFX; Bishop et al., 2009) and heparin (F; PDB 4C4N; Whalen et al., 2013) reveal overlapping interfaces that would prevent simultaneous binding. Binding footprints for each protein on SHH are shown below the corresponding structure, with hydrophilic interactions in pink and hydrophobic interactions in brown.
Mechanism of PTCH-mediated transport
Evidence of a shared mechanism with RND pumps comes from the observation that acidic residues that are required for proton flux and coupled transport in RND family proteins are functionally conserved in PTCH (Taipale et al., 2002). Unlike the bacterial membrane, there is no proton gradient across the membrane of animal cells; however, recent studies have suggested a requirement for the ubiquitous sodium gradient across the plasma membrane (Myers et al., 2017). Interestingly, DISP1 also belongs to the RND family and exports HH ligands by binding to their cholesteroyl appendages and then transferring them to Scube proteins, a similar mechanism (but opposite in direction) to the hand-off from the cholesterol carrier NPC2 to NPC1 (Creanga et al., 2012; Ma et al., 2002; Tukachinsky et al., 2012). Taken together, these observations suggest that PTCH may use a TM cation gradient to power the transport of sterols, presumably to influence SMO activity. Two functional studies support this idea. First, efflux of bodipy cholesterol from cultured fibroblasts is reduced by SHH, which blocks PTCH activity (Bidet et al., 2011). Second, overexpression of PTCH reduces cholesterol accessibility in the inner leaflet of the plasma membrane to a protein probe, which suggests that PTCH decreases either the abundance or chemical potential of inner leaflet cholesterol (Zhang et al., 2018).
The recently published PTCH structures suggest transport mechanisms. The structures note the presence of extra cryo-EM density, consistent with a ligand, in two places – a hydrophobic pocket within ECD1 and a V-shaped cavity adjacent to the SSD and facing the outer leaflet of the plasma membrane (Fig. 2A) (Gong et al., 2018). Although these electron densities are consistent with a sterol-like molecule, the resolution is insufficient for conclusive identification. The ECD1 and SSD sites are connected by a potential ‘tunnel’ (Fig. 2A) that could form a conduit for sterol movement through the protein during a transport cycle. Sterols could move in either direction through this tunnel – from the outer leaflet of the membrane to the ECD and ultimately to an acceptor (the direction of transport catalyzed by RND proteins) or from an extracellular donor through the ECD and down to the membrane (the direction of transport catalyzed by NPC1).
A lower resolution cryo-EM structure of PTCH carrying mutations in both ligand-binding sites is much more flexible than the ligand-bound structures and reveals a potential conformational change, a concerted ‘twisting’ motion that might be part of the transport cycle (Gong et al., 2018). Recent structures of the RND-family transporter, HpnN, show a similar ‘rigid-body swinging’ motion that may drive transport of bacterial lipids called hopanoids (structural and functional analogs of sterols) through a tunnel linking the periplasmic domain to the outer leaflet of the plasma membrane (Kumar et al., 2017). A prescient study published a decade ago (Hausmann et al., 2009) used evolutionary analysis to suggest that hopanoid transporters are the ancestors of PTCH and that the HH pathway might have evolved by co-opting parts of an ancient hopanoid sensing and transport pathway. These authors postulate that PTCH might inhibit HH signaling by locally depleting a hopanoid-like sterol that activates SMO, now a leading model for how PTCH regulates SMO (discussed below).
In evaluating the PTCH structures, it is important to remember that they are static snapshots that can only suggest models for PTCH function, which must be demonstrated experimentally. There are long-standing observations in the literature that are difficult to reconcile with these structures. For example, a mutant of PTCH that cannot bind and respond to SHH (Briscoe et al., 2001), called PTCHΔLoop2, lacks the entire ECD2 but is fully capable of inhibiting SMO, suggesting that the integrity of the proposed tunnel may not be essential (Fig. 2A). Second, the structures do not resolve the question of whether PTCH functions as an oligomer, similar to trimeric RND transporters (Lu et al., 2006).
How HH ligands inhibit PTCH
In addition to supporting a transporter-like function for PTCH, three of the recent structures show how HH ligands inhibit PTCH. The first two studies show two different interfaces between PTCH and SHH (Gong et al., 2018; Qi et al., 2018a). A third structure, determined under physiological calcium concentrations with a dually lipidated SHH ligand, reveals a complex with a 1SHH:2PTCH stoichiometry (Qi et al., 2018b). Here, a single SHH molecule engages two PTCH molecules using different interfaces (Fig. 2B). The first interface is between ECD1 of PTCH and the calcium- and zinc-binding surface of SHH (PTCH1-mol1 in Fig. 2B). At the second interface, the N-terminal palmitoyl group and subsequent 15 amino acids of SHH are inserted into the protein core of PTCH, and the C-terminal cholesteroyl moiety of SHH is inserted into ECD1 (Qi et al., 2018b; Qian et al., 2018). Both SHH-attached lipids occlude the putative tunnel that connects the ECD1 and SSD sterol binding sites (PTCH1-mol2 in Fig. 2B). Mutations that impair PTCH function can also decrease SHH binding to PTCH, consistent with the expectation that PTCH cycles through multiple conformations during its transport cycle, and that SHH selectively binds to and stabilizes one of these conformations (Gong et al., 2018; Tukachinsky et al., 2016).
Earlier biochemical studies predict this bipartite interaction between PTCH and SHH (Pepinsky et al., 1998; Taylor et al., 2001; Tukachinsky et al., 2016). The binding of SHH to PTCH has two consequences: it inhibits the transporter function of PTCH and it leads to its endocytosis and subsequent degradation (Incardona et al., 2002). Palmitoylation, which is not required for the high-affinity binding of SHH to PTCH, dramatically increases the signaling potency of SHH in vitro, indicating that PTCH binding and biochemical inactivation are separable events (Pepinsky et al., 1998). A palmitoylated 22 amino-acid N-terminal peptide of SHH (Palm-SHH22) is sufficient to inhibit PTCH function at high (micromolar) concentrations, likely through the interaction revealed by the SHH:PTCH1-mol2 structure (Fig. 2B) (Tukachinsky et al., 2016). However, unlike intact SHH, Palm-SHH22 cannot trigger PTCH endocytosis and degradation. Conversely, SHH lacking its N–terminal nine amino acids and the palmitate (SHHΔ9) fails to inactivate PTCH, but can still bind PTCH with high affinity and induce its endocytosis and degradation, likely through the interaction in the SHH:PTCH1-mol1 structure (Fig. 2B) (Tukachinsky et al., 2016; Williams et al., 1999). Interestingly, SHH lacking a palmitate (but including its N-terminal nine amino acids) can activate HH signaling both in vitro and in vivo, albeit with lower potency, suggesting that the conserved N-terminal nine amino acids of SHH may play a role in PTCH inactivation even without the palmitate modification (Chen et al., 2004; Pepinsky et al., 1998). In conclusion, the two different interfaces seen in the 1SHH:2PTCH structure may reflect the two different functions of PTCH: first, SMO inhibition regulated by the palmitate-based interface (PTCH1-mol2 in Fig. 2B), and second, ligand sequestration regulated by the protein-based interface (PTCH1-mol1 in Fig. 2B).
The interaction of HH ligands with co-receptors and antagonists
A conundrum raised by the SHH:PTCH1-mol1 structure is that the interaction interface between PTCH and SHH overlaps with the interface between SHH and several other cell-surface proteins and glycosaminoglycan chains of HSPGs, all of which are implicated in regulating ligand reception in target cells (Fig. 2C-F) (Bishop et al., 2009; Bosanac et al., 2009; Kavran et al., 2010; McLellan et al., 2008; Whalen et al., 2013). Vertebrate HH ligand co-receptors include three partially redundant proteins: the TM proteins CDO and BOC, and the glycosylphosphatidylinositol (GPI)-anchored protein GAS1. Elimination of all three proteins results in reduced HH signaling (Allen et al., 2011; Izzi et al., 2011). Although the binding of GAS1 to HH ligands is not well defined, it is clear that SHH cannot simultaneously interact with CDO/BOC and PTCH via the protein-based interface seen in the SHH:PTCH1-mol1 structure (Fig. 2C,D). However, the interaction between CDO/BOC and SHH (Fig. 2D) would leave the palmitate of SHH free to inactivate a molecule of PTCH, suggesting the possibility of a CDO/BOC-SHH-PTCH signaling complex, analogous to the PTCH-SHH-PTCH complex (Fig. 2B) (Qi et al., 2018b). Here, SHH could inactivate the biochemical function of PTCH without inducing its endocytosis and degradation. Alternatively, CDO and BOC might increase the local concentration of SHH on the cell surface and indirectly increase the chance of a SHH-PTCH interaction. Finally, HHIP is a secreted antagonist of HH ligands that binds to the same interface of SHH as both PTCH and CDO/BOC with low nanomolar affinity (Fig. 2E) (Chuang and McMahon, 1999). HHIP associates with the cell surface and extracellular matrix by binding to HSPGs (Holtz et al., 2015). An interesting question is why are CDO and BOC ligand agonists whereas HHIP is a ligand antagonist, even though both proteins bind to the same surface of SHH? Perhaps the higher affinity of HHIP for SHH, and its lack of tethering close to the plasma membrane, allow it to purely sequester extracellular HH ligands. In summary, co-receptors play key roles in regulating the availability of HH ligands and their influence on PTCH biochemical activity and PTCH trafficking.
Regulation of SMO
The endogenous ligand for SMO
Whereas HH ligands are received by PTCH and co-receptors, the HH signal is transmitted across the membrane by SMO, which belongs to the family of class F GPCRs, named for the frizzled (FZD) family of receptors for WNT ligands. SMO is composed of a heptahelical TMD, an extracellular cysteine-rich domain (CRD) and a linker domain (LD) that connects the CRD and TMD (Fig. 3A). Finally, SMO has an ∼240 amino acid C-terminal cytoplasmic tail domain (CCT) that is required for SMO localization to cilia and to activate downstream signaling, but is removed from all the proteins used for structural studies because of its partially disordered nature (Varjosalo et al., 2006).
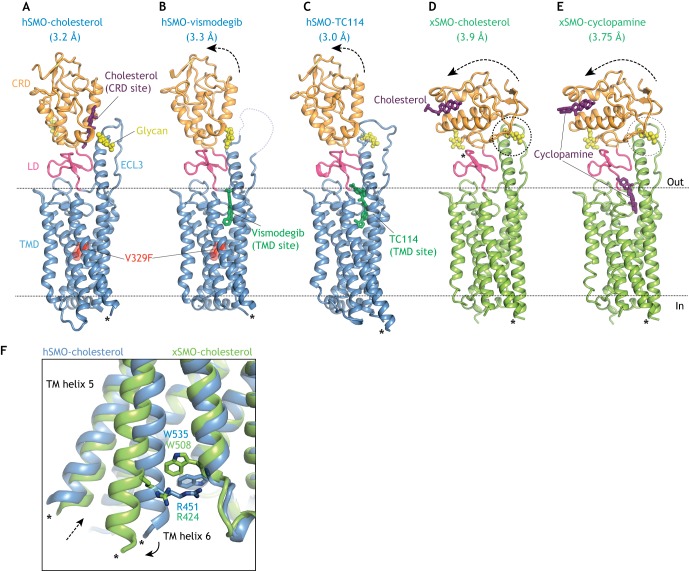
Fig. 3.
Multi-domain structures of SMO bound to agonists and antagonists. (A,B) Human SMO (hSMO, blue) carrying an inactivating V329F mutation in the TMD bound to the agonist cholesterol (A; PDB 5L7D) in the CRD-binding site or to the antagonist vismodegib (B; PBD 5L7I) in the TMD site (Byrne et al., 2016). (C) hSMO bound to the TMD antagonist TC114 (PBD 5V57; Zhang et al., 2017). Arrows show movement of the CRD in the antagonist-bound structures (B,C) relative to the cholesterol bound structure (A). (D,E) Structures of Xenopus SMO (xSMO, green) bound to the agonist cholesterol (D; PDB 6D35; Huang et al., 2018) or the antagonist cyclopamine (E; PDB 6D32) are identical and show a dramatic re-orientation of the CRD relative to the TMD. Dotted circles (D,E) highlight a potential steric clash between the CRD and an N-linked glycan in the third extracellular loop of SMO. The N-linked glycans for Xenopus SMO were modeled because they were removed for crystallization. (F) Overlay of the indicated SMO structures showing rupture of the ionic lock between a tryptophan (W) and an arginine (R) residue resulting in the outward movement of TM6 (solid arrow) and the opening to a hydrophobic channel proposed to run through the xSMO TMD in the activated state (dotted arrow). Asterisks in all structures show the connections to the BRIL or flavodoxin domains interposed between TM5 and TM6 to facilitate crystallization. CRD, cysteine-rich domain; ECL3, extracellular loop 3; LD, linker domain; TMD, transmembrane domain.
Three observations led to the hypothesis that PTCH regulates a small molecule ligand for SMO: the similarity of PTCH to transporter-like proteins described above, the lack of physical interactions between PTCH and SMO, and the observation that each molecule of PTCH can inhibit multiple molecules of SMO (Denef et al., 2000; Ingham et al., 2000; Taipale et al., 2002). Several structures of SMO have been determined by X-ray crystallography, both of the isolated TMD and CRD and of the multi-domain CRD-TMD (Byrne et al., 2016; Huang et al., 2016, 2018; Nachtergaele et al., 2013; Rana et al., 2013; Wang et al., 2013, 2014; Zhang et al., 2017). The structures provide an atomic view of two major ligand-binding sites in SMO: the CRD site and the TMD site (Fig. 3A,B). A number of synthetic and natural small molecules bind to both these sites to positively or negatively regulate SMO activity (Fig. 3A-E). We refer the reader to reviews on small molecule regulation of SMO for a more detailed discussion (Byrne et al., 2018; Sharpe et al., 2015). We focus below on the endogenous ligand that mediates communication between PTCH and SMO in vertebrates.
Several different approaches suggest that the endogenous SMO regulator is a sterol lipid. In addition to the homology of PTCH to NPC1, an early indication came from the discovery that a plant sterol alkaloid cyclopamine binds to and inhibits SMO at the TMD site (Chen et al., 2002a; Taipale et al., 2000), which has since become the target of pharmaceutical SMO inhibitors (Chen et al., 2002b; Frank-Kamenetsky et al., 2002). Second, pharmacological or genetic approaches that reduce cellular cholesterol levels also attenuate HH signaling in target cells, showing that cholesterol plays a second role in signal reception distinct from its role in ligand biogenesis (Blassberg et al., 2016; Cooper et al., 1998, 2003; Incardona and Roelink, 2000). Third, side-chain oxysterols (endogenous metabolites of cholesterol) activate HH signaling at the level of SMO and induce its accumulation in primary cilia – even in the absence of HH ligands (Corcoran and Scott, 2006; Dwyer et al., 2007; Rohatgi et al., 2007). Pharmacological and ligand-affinity studies led to the discovery of a second oxysterol-binding site in SMO that is entirely distinct from the TMD-binding site (Nachtergaele et al., 2012). This second site was later shown to be formed by a shallow hydrophobic groove in the CRD site – the same groove in FZD receptors that binds to the palmitoleyl modification on WNT ligands (Fig. 3A) (Janda et al., 2012; Myers et al., 2013; Nachtergaele et al., 2013; Nedelcu et al., 2013). Though physically separate, the oxysterol-binding site in the CRD is allosterically linked to the TMD-binding site (Nachtergaele et al., 2012). Interestingly, cyclopamine, although initially characterized as a TMD antagonist, can also function as an agonist by binding the CRD site (Huang et al., 2016; Nachtergaele et al., 2013) (Fig. 3E), though its dominant effect on signaling in cells is inhibitory. Recent studies suggest that endogenous oxysterols enriched in the ciliary membrane may activate SMO through the CRD site in specific developmental or oncogenic contexts (Raleigh et al., 2018).
The first multi-domain structure of SMO unexpectedly reveals a cholesterol molecule bound to the CRD site in the same position that oxysterols were predicted to bind, raising the possibility that the endogenous ligand for the CRD is cholesterol itself, rather than oxysterols (Byrne et al., 2016) (Fig. 3A). Mutations in the CRD site that prevent cholesterol binding impair HH signaling in cultured cells (Byrne et al., 2016) and mouse embryos (Xiao et al., 2017). Two independent studies demonstrate that cholesterol is sufficient to activate signaling even in the absence of HH ligands (Huang et al., 2016; Luchetti et al., 2016). Both studies identify mutations in the CRD that could discriminate between cholesterol-activation and oxysterol-activation, and show that mutations that prevent oxysterol activation (but leave cholesterol activation intact) have little effect on SHH-induced signaling. Low doses of cholesterol (but not oxysterols) synergize with SHH in signaling assays (Huang et al., 2016; Luchetti et al., 2016; Nachtergaele et al., 2012). Collectively, these results suggest that cholesterol itself may be the endogenous small molecule SMO agonist regulated by PTCH – the elusive second messenger that communicates the HH signal between its receptor and the TM transducer (Byrne et al., 2016; Huang et al., 2016; Luchetti et al., 2016). The model proposed by these studies provides a unifying explanation for the diverse set of observations made over nearly two decades that link cholesterol to the reception of HH signals.
The mechanism of SMO activation
Although these studies point to a role for cholesterol in the PTCH-SMO interaction, there is uncertainty about which domain of SMO is targeted by the inhibitory effect of PTCH. Complete deletion of the CRD (in SMOΔCRD), or mutations that destabilize the CRD-TMD interface, increase the constitutive (ligand-independent) signaling activity of SMO in cells (Byrne et al., 2016). SMOΔCRD is markedly less sensitive to PTCH, but its activity can be suppressed by PTCH overexpression (Myers et al., 2013). One possibility is that PTCH regulates the access of both the CRD and TMD to cholesterol. Cholesterol is not bound to the TMD in any of the solved SMO structures, even though SMO was crystallized with a high concentration of cholesterol. The classical TMD site (Fig. 3B) is unlikely to mediate the effect of cholesterol because various mutations in this site fail to alter SHH-driven signaling (Dijkgraaf et al., 2011; Myers et al., 2013). However, molecular dynamic simulations identify a potential cholesterol-binding site between the extracellular ends of TM2 and TM3, at the outer leaflet of the plasma membrane (Fig. 4A) (Hedger et al., 2018). Furthermore, mutagenesis and computational docking studies identify a putative oxysterol-binding site at the cytoplasmic end of the TMD (Fig. 4A) (Raleigh et al., 2018). Finally, a recent multi-domain structure of SMO suggests that cholesterol from the inner leaflet of the plasma membrane could gain access to a hydrophobic tunnel in the center of the TMD bundle through a gate at the cytoplasmic end of the TMD (Fig. 3F) (Huang et al., 2018). It will be important to determine whether any of these proposed SMO-sterol interactions can explain the ability of PTCH to inhibit the activity of SMOΔCRD.
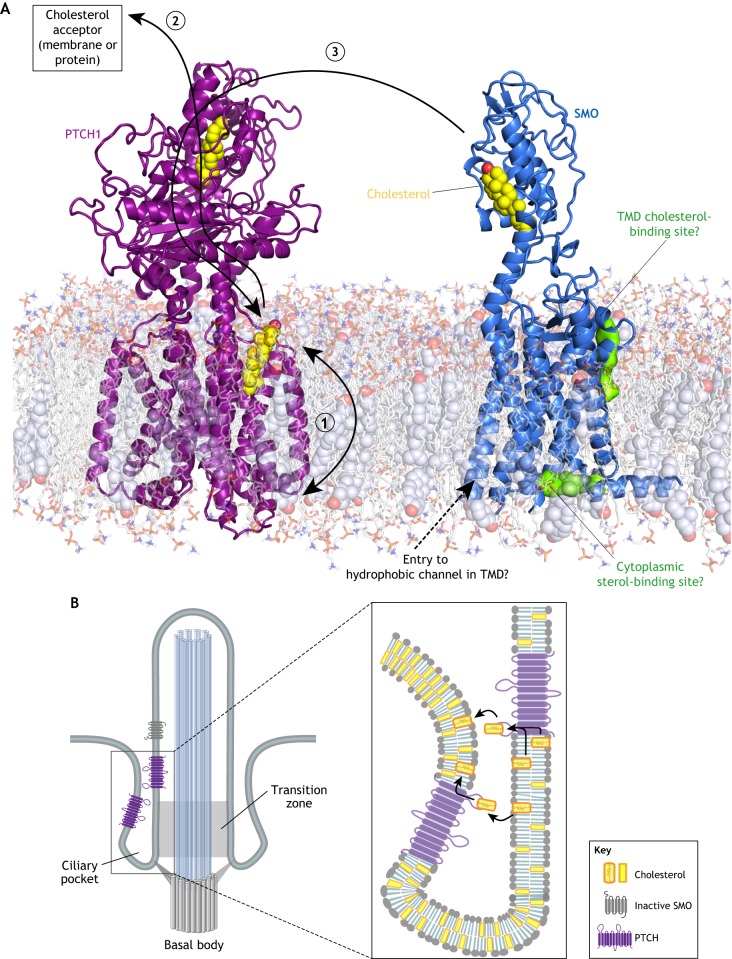
Fig. 4.
Models for how PTCH inhibits SMO. (A) Schematic of PTCH and SMO embedded in a model lipid bilayer. Structurally identified cholesterol molecules bound to PTCH and SMO are depicted as yellow spheres. Two potential sterol-binding sites on SMO identified by computational methods are shown as green surfaces. Three possible sterol transport paths are shown by black arrows. In model 1, PTCH reduces the abundance or accessibility of inner leaflet cholesterol, preventing it from interacting with the hydrophobic channel or the cytoplasmic sterol-binding site of SMO. In model 2, cholesterol moves through PTCH from the outer leaflet of the membrane to the ECD1 and eventually to a protein or membrane acceptor, thereby depleting the membrane of cholesterol. In model 3, PTCH accepts cholesterol from the SMO CRD (or another donor) and transports it to the membrane, thereby turning off SMO activity. (B) Models for how PTCH could deplete cholesterol from the ciliary membrane (thereby reducing its access to SMO) by transporting it between the two closely opposed membranes of the ciliary pocket.
Transmission of the HH signal across the membrane requires a conformational change in the TMD of SMO, so there is considerable interest in understanding the structural transitions that are associated with activation. Interactions between the CRD and the TMD play a key role in stabilizing the inactive state of SMO; mutations that destabilize the linker domain that connects the CRD to TMD or that introduce glycosylation sites at the CRD-TMD interface increase the constitutive activity of SMO (Byrne et al., 2016). Small angle X-ray scattering (SAXS) shows that activation involves the movement of the CRD relative to the TMD (Byrne et al., 2016). However, the precise orientation of the CRD relative to the TMD in active SMO remains uncertain (Fig. 3A-E and Box 2). One of the putative active-state structures highlights a cation-pi bond (the ‘ionic lock’) between an arginine and a tryptophan residue at the cytoplasmic end of the TM bundle (Huang et al., 2018) (Fig. 3F). This ionic lock is broken by known oncogenic mutations that activate SMO and is also conserved in the FZD group of WNT receptors (Wright et al., 2019). SMO activation may result in the rupture of this ionic lock, leading to the outward movement of TM6 and the consequent exposure of a new molecular surface to engage a cytoplasmic effector.
Box 2. Structural models of SMO activation.
All of the multi-domain SMO structures described to date, regardless of whether they are presented as active- or inactive-state conformations, are derived from proteins that lack signaling capacity. To enable crystallization, mutations or small molecules are used to stabilize the inactive state and heterologous protein domains inserted to facilitate crystal contacts. One model for activation proposes that the CRD moves away from a long helical extension formed by the third extracellular loop to allow cholesterol binding (Fig. 3A,B) (Byrne et al., 2016). However, this model is based on the structure of SMO carrying a mutation in the TMD that stabilizes the inactive state and so does not provide insights into TMD conformational changes (Fig. 3A,B). A second study proposes a conformational change in the CRD itself that is propagated to the TMD (Huang et al., 2016); however, this conformation is unlikely to be physiological as it is induced by a zinc-promoted crystal contact (discussed by Luchetti et al., 2016). A third structural model (Fig. 3D,E) shows a large reorientation of the CRD relative to the TMD (Huang et al., 2018); however, this active-state conformation would produce a steric clash with N-linked glycans removed for determining the structure (dotted circle in Fig. 3D,E) and thus is unlikely to be adopted by endogenous SMO in cells. In addition, the TMD conformations of the cholesterol-bound and cyclopamine-bound structures are identical (Fig. 3D,E), even though we know that cyclopamine binding to the TMD inhibits SMO signaling. Overall, none of the structures fully explains the difference between active- and inactive-state SMO. Additional active-state structures for SMO, ideally in complex with a downstream effector, are required to resolve the details of the structural changes that drive signal transmission across the membrane.
How PTCH inhibits SMO
The recent flurry of PTCH and SMO structures allow informed speculation on how PTCH might prevent SMO access to cholesterol (Fig. 4A). One possibility is that PTCH alters the trans-bilayer distribution of cholesterol in the plasma membrane (Fig. 4A, model 1) (Zhang et al., 2018). By reducing cholesterol abundance or accessibility in the inner leaflet of the plasma membrane, PTCH could prevent cholesterol access to the proposed tunnel in SMO TMD (Huang et al., 2018) or to the sterol-binding site proposed on the cytoplasmic end of the TMD (Fig. 4A) (Raleigh et al., 2018). Alternatively, by reducing outer leaflet cholesterol, PTCH could prevent cholesterol access to the computationally predicted site at the extracellular end of the TMD or even to the SMO CRD – if indeed the CRD picks up the cholesterol from the outer leaflet (Hedger et al., 2018). However, it is not clear that significant differences between inner and outer leaflet cholesterol abundance can be sustained given the rapid flip-flop rate of cholesterol in membranes (Steck and Lange, 2018). The second possibility is that PTCH pumps cholesterol from its SSD to its ECD1 (and eventually to an unidentified protein or membrane acceptor), thereby depleting both the outer and inner leaflets of cholesterol (Gong et al., 2018; Qi et al., 2018b) (Fig. 4A, model 2). Finally, analogous to the way NPC1 accepts cholesterol from NPC2 and transfers it to the membrane, PTCH could inactivate SMO by catalyzing cholesterol transfer from the SMO CRD to the membrane, perhaps again through the tunnel that connects the ECD1 to the SSD (Fig. 4A, model 3).
How can PTCH prevent SMO access to cholesterol when cholesterol constitutes ∼30% of lipid molecules in the plasma membrane (Steck and Lange, 2018)? A potential solution to this vexing question is that PTCH operates in a membrane compartment that is segregated from the large cholesterol pool in the bulk plasma membrane (Fig. 4B). The primary cilium has been proposed to be this privileged compartment (Huang et al., 2016; Luchetti et al., 2016), as PTCH localizes in the ciliary membrane and in membranes around the base of the cilium both in vitro and in vivo (Rohatgi et al., 2007). The base of the cilium, where PTCH staining is most prominent in HH-responsive embryonic tissues (Rohatgi et al., 2007), includes the transition zone and is encircled by the ciliary pocket, formed when the ciliary membrane folds back on itself (Fig. 4B) (Rohatgi and Snell, 2010). PTCH could control ciliary cholesterol by transporting it between the two closely opposed membranes of the ciliary pocket (Fig. 4B). Interestingly, the cilium is the only membrane-bound compartment in the cell where PTCH and SMO are both localized simultaneously (Rohatgi et al., 2009). In summary, PTCH may inhibit SMO by using its transporter-like activity to reduce the abundance of accessible cholesterol in the membrane of the cilium or the ciliary pocket. However, we emphasize that this model requires significant additional experimental testing; it remains possible that PTCH regulates SMO through a different sterol lipid that is less abundant than cholesterol.
Signal transmission from SMO
In contrast to the Drosophila pathway, the issue of how vertebrate SMO transmits the HH signal from the cell membrane to the cytoplasm remains unresolved, with multiple implicated components. One important point to remember is that just because a component is required for signaling, it does not mean that its activity is changed by SMO signaling. A change in biochemical activity in response to HH ligands has not been conclusively demonstrated for any of the myriad components (many with strong loss-of-function phenotypes) implicated in SMO signaling to the cytoplasm. Our focus below is on signaling mechanisms that relay signals from SMO to the GLI transcription factors, but we note that SMO can activate other non-transcriptional signaling outputs (Box 3).
Box 3. GLI-independent mechanisms of SMO signaling.
SMO regulates several cellular processes independent of GLI proteins. In the nervous system, SMO signaling mediates both attractive and repulsive responses during axon guidance and calcium spiking in the developing spinal cord (Belgacem and Borodinsky, 2011; Charron et al., 2003; Yam et al., 2009). SMO can regulate cytoskeletal responses through the small GTPases RhoA and Rac1 (Chinchilla et al., 2010) and metabolic reprogramming through a calcium-AMPK pathway (Teperino et al., 2012). Although the signaling mechanisms in each case remain to be fully elucidated, they frequently involve activation of the Gαi family of heterotrimeric G-proteins.
In all animals, a central task of SMO is to antagonize the inhibitory effect of PKA, a multifunctional kinase, on the GLI family of transcription factors (Fig. 1A). In principle, SMO could regulate PKA in one of two general ways, although the challenge in both cases is how to selectively inhibit the effects of PKA on the HH pathway, while sparing any effect on non-HH PKA substrates in the cell. The first possibility is that active SMO reduces PKA enzymatic activity in a specific subcellular compartment relevant to HH signaling (such as the primary cilium) or in a HH-specific protein complex. Alternatively, SMO could reduce the access of GLI proteins to PKA by segregating GLI and PKA in different compartments or different complexes.
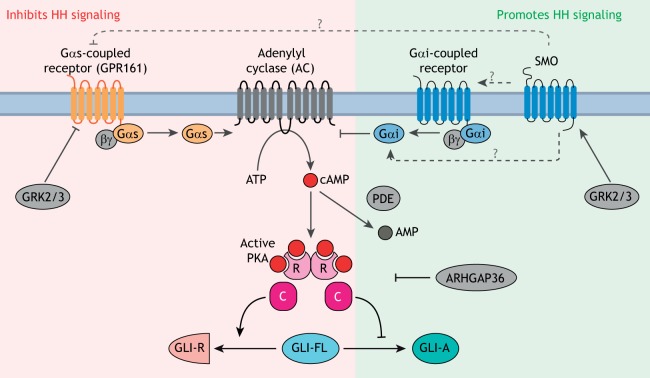
PKA activity can be increased or decreased by a signaling cascade initiated by GPCRs (Fig. 5). When GPCRs are activated by ligands, they catalyze the exchange of guanosine diphosphate (GDP) for guanosine triphosphate (GTP) on the Gα subunit of heterotrimeric G-proteins, resulting in their activation and dissociation from the Gβ and Gγ subunits. Gαs subunits activate adenylyl cyclases (AC), which synthesize the second messenger cyclic adenosine 3′,5′-monophosphate (cAMP) from ATP; Gαi subunits reduce cAMP levels by inhibiting AC (Fig. 5). cAMP directly binds and activates PKA. The balance of Gαi and Gαs activities, along with the activities of phosphodiesterases (PDEs) that hydrolyze cAMP, sets the cellular cAMP concentration and hence the PKA activity level.
Fig. 5.
GLI proteins are regulated by PKA. PKA is a conserved inhibitor of GLI proteins, and the strength of HH signaling is inversely correlated with the activity of PKA in cells. The pathway regulating cAMP levels and PKA activity in cells is shown: proteins that increase PKA activity (red background) inhibit HH signaling, whereas those that decrease PKA activity (green background) enhance HH signaling. Among positive regulators of signaling, GPCRs coupled to Gαi reduce cAMP synthesis by inhibiting AC, phosphodiesterases (PDEs) hydrolyze cAMP and ARHGAP36 inhibits PKA. GPCRs coupled to Gαs, such as GPR161, inhibit signaling by increasing cAMP synthesis by AC. The kinases GRK2 and GRK3 are strong positive regulators that may function either by downregulating GPCRs coupled to Gαs or by directly promoting SMO activity. The mechanism by which SMO antagonizes the PKA axis is not clear (dashed arrows with ‘?’), but may involve direct activation of Gαi, inhibition of a Gαs-coupled GPCR or activation of a Gαi-coupled GPCR. PKA is composed of catalytic (C) and regulatory (R) subunits.
The relationship between HH signaling and PKA activity is well established: increasing PKA activity inhibits signaling, whereas decreasing PKA activates signaling (even in the absence of HH ligands) in a variety of systems ranging from cultured cells to vertebrate embryos. This is shown by directly manipulating PKA activity or the upstream ACs, PDEs and Gα proteins (Fig. 5) (Fan et al., 1995; Hammerschmidt et al., 1996; Huang et al., 2002; Hynes et al., 1995; Regard et al., 2013; Tuson et al., 2011; Vuolo et al., 2015; Wechsler-Reya and Scott, 1999; Williams et al., 2015; Yao and Capel, 2002).
Regulation of heterotrimeric G-proteins by SMO
Given the genealogy of SMO as a GPCR, the most parsimonious mechanism of SMO signaling would be through the activation of G-proteins (reviewed by Ayers and Thérond, 2010). SMO can activate the Gαi family of heterotrimeric G-proteins (encoded by the Gnai1, Gnai2, Gnai3, Gnao1 and Gnaz genes in mammals) in response to agonists (DeCamp et al., 2000; Riobo et al., 2006; Shen et al., 2013) (Box 3). As Gαi proteins inhibit AC and reduce cAMP levels, their activation would be predicted to reduce PKA activity and hence activate the GLI proteins (Fig. 5). However, in vertebrates, it is not clear that SMO regulates Gαi, AC and PKA activity during the course of endogenous signaling. Redundancy between multiple Gnai genes in vertebrates prevents clean loss-of-function studies. Instead, many studies use pertussis toxin (Ptx), which promotes uncoupling of all Gαi proteins (except Gαz) from upstream GPCRs. In culture, Ptx addition attenuates (but does not eliminate) SHH-driven reporter gene expression in fibroblasts (Low et al., 2008; Riobo et al., 2006). Zebrafish embryos injected with Ptx RNA exhibit a fusion of midline structures and reduction in HH target gene expression, consistent with reduced HH activity (Hammerschmidt and McMahon, 1998). However, subsequent attempts to suppress Gαi activity in developing vertebrate systems do not support a role in HH signaling. Electroporation of dominant negative Gαi2 or the S1 catalytic subunit of Ptx into the developing chicken neural tube failed to disrupt HH-sensitive progenitor domains (Low et al., 2008). Lastly, comprehensive elimination of Gαi function in the developing mouse limb through the conditional expression of the Ptx S1 catalytic subunit in a Gαz−/− background has no discernible effect on HH-dependent skeletal development or limb patterning (Regard et al., 2013).
Gαs, which is encoded by a single Gnas gene in vertebrates, is a negative regulator of HH signaling, consistent with its role in activating AC (Fig. 5). Disruption of Gnas in both the developing neural tube and in neural progenitor cell cultures results in elevated HH signaling activity and an expansion of HH-dependent cell types (Pusapati et al., 2018b; Regard et al., 2013). Similarly, the conditional loss of Gnas in the developing limb mesenchyme results in heterotopic ossification due to ectopic HH signaling (Regard et al., 2013). Finally, tissue-specific loss of Gnas can drive the development of medulloblastoma in the cerebellum (He et al., 2014b) and basal cell carcinoma in the skin (Iglesias-Bartolome et al., 2015), partially because of unrestrained activation of HH signaling. Despite the strong genetic evidence that Gαs restrains HH signaling, there is no conclusive evidence that SMO, either directly or indirectly, regulates Gαs activity during the course of endogenous HH signaling.
Regulation of other GPCRs by SMO
Rather than regulating Gα proteins directly, SMO could influence PKA activity by regulating other GPCRs. The best candidate is GPR161, a ciliary GPCR that activates Gαs and functions as a negative regulator of HH signaling in cells and embryos (Hwang et al., 2018; Mukhopadhyay et al., 2013) (Fig. 5). SMO activation and ciliary accumulation leads to the clearance of GPR161 from the ciliary membrane, which suggests that ciliary GPR161 elevates local PKA activity, thereby suppressing basal HH signaling (Mukhopadhyay et al., 2013). By clearing GPR161 from cilia, SMO activation would lead to a drop in ciliary PKA activity and consequent activation of HH signaling. However, although GPR161 clearly attenuates HH signaling, it is not a required component of signaling downstream of SMO. Gpr161−/− NIH/3T3 cells exhibit no discernible elevation in baseline HH signaling activity (Pusapati et al., 2018b). SHH and SMO agonists can activate HH signaling in Gpr161−/− NIH/3T3 cells at all doses, albeit with higher potency and efficacy than wild-type cells (Pusapati et al., 2018b). Consistent with its role in modifying signaling strength, the mouse embryonic phenotypes of GPR161 loss are much milder than the phenotypes of embryos lacking PKA or Gαs (Regard et al., 2013; Tuson et al., 2011).
Another role for GPCR regulation in SMO signaling is suggested by the strong positive regulation of HH signaling by GPCR kinase (GRK2) in both Drosophila and vertebrates (Chen et al., 2010; Meloni et al., 2006; Philipp et al., 2008). The positive role of GRK2 in HH signaling is opposite to its known role in attenuating GPCR signaling by direct receptor phosphorylation. Partial redundancy between Grk2 and Grk3 probably accounts for the relatively mild HH signaling phenotype in Grk2−/− mice, although this has not been tested by the analysis of double null embryos (Philipp et al., 2008; Pusapati et al., 2018b). Furthermore, inhibition of GRK2/3 in fibroblasts or neural progenitor cells completely blocks HH signaling (Pusapati et al., 2018b). The HH-specific target of GRK2/3 remains controversial and two models have been proposed. First, GRK2/3 directly phosphorylates SMO and facilitates its activation and accumulation in primary cilia (Chen et al., 2011). Second, GRK2/3 facilitates the clearance of GPR161 from cilia (Pal et al., 2016). However, SMO ciliary accumulation is not affected in zebrafish embryos lacking GRK2/3 activity and GRK2/3 is required for HH signaling even in the absence of GPR161 (Pusapati et al., 2018b; Zhao et al., 2016). Definitive identification of the GRK2 substrate, whether it is SMO itself or another unknown GPCR(s), will shed further light on the mechanism of SMO signaling.
Local regulation of PKA activity
If the regulated step in SMO signaling is the inhibition of PKA activity, one should be able to measure decreases in PKA enzymatic activity, decreases in cAMP concentrations or changes in Gα localization in response to HH ligands. Much of the focus in the literature has been on the primary cilium, with current models suggesting that SMO activation and accumulation in cilia leads to decreases in local cAMP levels and PKA activity, allowing GLI activation during its transit through the cilium (Fig. 1C) (Mukhopadhyay et al., 2013; Tuson et al., 2011). PKA regulatory and catalytic components have been localized at the centrosome and cilia base, and specific AC isoforms are concentrated in the ciliary membrane (Barzi et al., 2010; Mick et al., 2015; Tuson et al., 2011; Vuolo et al., 2015). However, Gαi and Gαs proteins have not been reproducibly detected in cilia, raising the question of how SMO would regulate local enzymatic activity of AC and PKA. In addition, there is a paucity of data showing any changes in either cAMP levels or PKA activity in response to SMO activation, either globally in the cell or locally in cilia. A study using fluorescence resonance energy transfer-based cilia-targeted biosensors shows that prolonged exposure to HH agonists reduces the concentration of cAMP and kinase activity of PKA in cilia (Moore et al., 2016). Interestingly, this effect is independent of Gαs or Gαi but instead depends on the direct inhibition of ACs by ciliary calcium influx induced by SMO. Further development of time-resolved tools to measure (and perturb) signaling components selectively at cilia is a promising approach to test whether changes in ciliary cAMP or PKA occur with kinetics consistent with an instructive role in HH signaling.
Much of the extensive literature on the effects of manipulating PKA, or the proteins that regulate PKA (AC, Gαi, Gαs, GPR161, PDE; see Fig. 5), on HH signaling activity is consistent with the view that HH signaling strength in target cells is exquisitely sensitive to the basal level of PKA activity (Humke et al., 2010). Hence, any manipulation that increases basal PKA activity in cells will inhibit HH signaling. Conversely, any manipulation that decreases PKA activity will enhance HH signaling strength or even induce ectopic signaling. However, these results do require that PKA, or any of the proteins that control its activity, are directly regulated by SMO. An alternative model is that SMO shields GLI proteins from the inhibitory influence of PKA. One possibility is that the ciliary trafficking of GLI proteins, regulated by SMO, may serve to regulate GLI access to PKA (Tukachinsky et al., 2010; Tuson et al., 2011).
The uncertainty around how SMO regulates PKA inhibition of GLI proteins can be resolved by conclusively identifying the downstream target of active SMO in vertebrates. In the absence of redundancy or compensation, the effect of disrupting such a target should be just as strong as that of disrupting SMO. Although targeted proteomic studies have failed in this endeavor (likely because of the transient or detergent-sensitive nature of the interaction), the application of proximity biotinylation-based proteomics, which can identify protein interactions in intact cells, or unbiased CRISPR-based genetic screens are promising new approaches that may soon resolve this long-standing mystery.
Regulation of the GLI transcription factors by a multi-site phosphorylation code
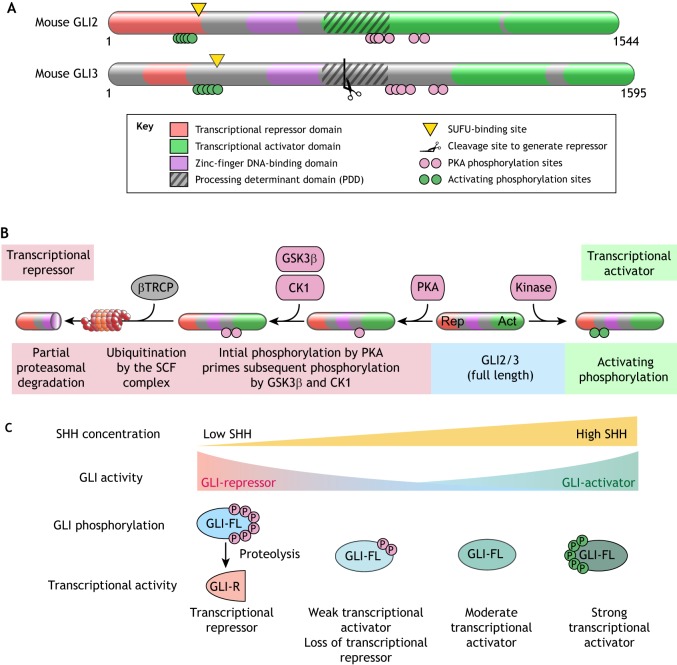
HH signaling converges on a small family of zinc-finger transcription factors encoded by Gli genes (Gli1, Gli2 and Gli3) in vertebrates (Fig. 6A) (Hui et al., 1994). In vertebrates, GLI2 and GLI3 proteins (together referred to as GLI2/3) can exist in at least three states: proteolytically processed transcriptional repressors (GLI2/3R), full-length transcriptionally inactive proteins (GLI2/3FL), and full-length transcriptional activators (GLI2/3A) (Fig. 6B) (reviewed by Hui and Angers, 2011; see also Aza-Blanc et al., 1997; Methot and Basler, 1999; Wang et al., 2000). Mouse genetic studies have shown that the transcriptional activator function is largely allocated to GLI2 and transcriptional repressor function to GLI3 (Litingtung and Chiang, 2000; Matise et al., 1998). GLI1 is a transcriptional target of GLI2 and GLI3, and functions exclusively as a transcriptional activator that amplifies existing HH signaling activity. We will focus our discussion on GLI2/3 below, as they are directly regulated by SMO signaling. GLI1 is regulated by distinct mechanisms, especially important for HH-driven cancers (Atwood et al., 2013; Huntzicker et al., 2006; Mirza et al., 2019).
Fig. 6.
Regulation of GLI proteins in HH signaling. (A) Domain structures of mouse GLI2 and GLI3 proteins. Regions of the protein involved in various biochemical reactions are annotated. (B) Sequence of reactions that convert full-length GLI2/3 (blue shading) into a transcriptional repressor (red shading) or a transcriptional activator (green shading). Phosphorylation by PKA primes further phosphorylation by GSK3β and CK1 and promotes recognition by βTRCP, the substrate recognition adaptor of the SCF E3 ubiquitin ligase (Tempe et al., 2006; Wang and Li, 2006). SCFβTRCP-mediated ubiquitination targets GLI2/3 to the proteasome for degradation. Owing to the presence of a processing determinant domain (PDD, see A), GLI3 (and to a lesser extent GLI2) are subject to an unusual partial proteasomal degradation reaction that generates fragments that function as pure transcriptional repressors (Schrader et al., 2011). (C) Multiple states of GLI activity can be encoded by different patterns of GLI phosphorylation. Full phosphorylation of GLI at the PKA phosphorylation sites (red circles) drives proteolytic production of GLI-R and repression of target genes. Graded dephosphorylation at the PKA sites prevents formation of GLIR and increases the ability of GLI to activate transcription. Maximum GLI transcriptional activity is associated with a separate activating hyper-phosphorylation (green circles).
How are the functions of primary cilia, SUFU and PKA coordinated by HH ligands to produce the multiple states of GLI2/3 activity that are required for graded responses? Epistasis experiments show that primary cilia appear to function at a step between PKA and SUFU – the loss of primary cilia blocks signaling that is triggered by PKA inhibition but has no effect on signaling that is triggered by loss of SUFU (Chen et al., 2009; Ocbina and Anderson, 2008). GLI2/3, in association with SUFU, travel through primary cilia both in the absence and presence of HH ligands, though their abundances at the tips of cilia increase in response to SMO activation (Haycraft et al., 2005; Kim et al., 2009; Tukachinsky et al., 2010) (Fig. 1C). The consequences of GLI-SUFU trafficking through cilia is entirely different in the absence and presence of SMO activity (Humke et al., 2010; Tukachinsky et al., 2010; Tuson et al., 2011) (Fig. 1C). When SMO is inactive, ciliary trafficking promotes the partial proteolytic processing of GLI2/3FL into GLI2/3R fragments that dissociate from SUFU and enter the nucleus to repress target genes (Figs 1C and 6B). When SMO is active and accumulates in cilia, ciliary trafficking promotes the dissociation of SUFU from full-length GLI2/3, allowing the formation of GLI2/3A proteins that can enter the nucleus and activate target genes (Fig. 1C). The biochemical mechanisms that link cilia trafficking to either GLI2/3 proteolysis or to SUFU dissociation from GLI2/3 remain largely obscure. However, they are likely controlled by the phosphorylation of GLI2/3 by PKA (which in turn is regulated by the activity-state of SMO) and by the atypical kinesin KIF7 (Fig. 1C) (reviewed by Hui and Angers, 2011).
Regulation of the GLI2/3 activity state by PKA is key for graded HH ligand responses to be converted into multiple levels of GLI activity in the nucleus, and ultimately to alternative differentiation outcomes in target cells (Stamataki et al., 2005). Multi-site phosphorylation of the GLI2/3 proteins is used to resolve these graded responses into multiple discrete activity states, referred to as multi-stability (Fig. 6C) (Thomson and Gunawardena, 2009). There is evidence, both from cultured cells and chicken neural tube development, that the transcriptional activity of GLI2/3 is regulated by a multi-site phosphorylation code that is inscribed on two clusters of phosphorylation sites (Fig. 6A) (Niewiadomski et al., 2014). Although the PKA sites were originally thought to only control proteolysis to generate the GLI2/3R fragment (Fig. 6B), more recent mutagenesis and phosphorylation analysis using mass spectrometry shows that increasing stoichiometry of phosphorylation at the PKA-target sites can inhibit the transcriptional activity of full-length GLI2/3 proteins in a graded fashion (Niewiadomski et al., 2014) (Fig. 6C). Notably in Drosophila, multi-site phosphorylation of both SMO and the KIF7 ortholog Cos2 has been implicated in enabling graded signaling responses, suggesting a common regulatory device to encode multistability in morphogen signaling systems (Ranieri et al., 2012; Su et al., 2011).
Mechanisms that regulate target cell sensitivity to HH ligands
Although ligand secretion and distribution play important roles in regulating the strength of HH signaling, more recent work has uncovered signaling mechanisms that tune the sensitivity of target cells to HH ligands. GPCRs can modify target-cell sensitivity to HH ligands, presumably by changing PKA activity (Fig. 5). The best studied of these is GPR161 (described above), which reduces the sensitivity of target cells to HH ligands, likely by activating Gαs and consequently PKA activity (Mukhopadhyay et al., 2013; Pusapati et al., 2018b). GPR161 has been shown to attenuate HH signaling in the developing limb, skeleton and spinal cord, and deletion of GPR161 in neural stem cells can induce cerebellar tumors (Hwang et al., 2018; Mukhopadhyay et al., 2013; Shimada et al., 2018). A ligand that regulates GPR161, positively or negatively, remains to be identified. In the cerebellum, the GPCR ADCYAP1R1 attenuates HH signaling, both in the context of normal cerebellar development and HH-driven medulloblastoma, by increasing PKA activity through Gαs (Niewiadomski et al., 2013). PKA can also be regulated by mechanisms other than GPCRs to influence HH signaling. ARHGAP36, a RHO-GTPase activating protein that is overexpressed in HH-driven medulloblastoma, potentiates HH signaling by both inhibiting PKA enzymatic activity and inducing PKA degradation (Eccles et al., 2016; Rack et al., 2014). Neuropilins are TM receptors that positively regulate signaling by activating PDE4D to induce the hydrolysis of cAMP (Ge et al., 2015) (Fig. 5).
A genome-wide CRISPR screen recently uncovered a set of three proteins that dampen target cell sensitivity to HH ligands by reducing the abundance of SMO on the cell surface and primary cilium (Pusapati et al., 2018a). Two of these proteins, MEGF8 and MOSMO, are TM proteins, and a third (MGRN1) is a RING-family E3 ubiquitin ligase, suggesting that this regulation may involve SMO ubiquitination and that it may be regulated by a yet-undiscovered ligand. Mouse and human phenotypes of MEGF8 mutations are consistent with tissue-specific roles in HH signaling (Twigg et al., 2012; Zhang et al., 2009), but elucidating the developmental and oncogenic roles of these proteins requires further genetic analysis. These findings raise the possibility that membrane protein trafficking events at the cell surface or the primary cilium might play roles in regulating target-cell responses to HH ligands.
We expect that these, and perhaps other undiscovered modules, will play important roles in modifying responses to HH ligands, thus allowing the same core signaling system to be adapted for use in myriad developmental, regenerative and oncogenic contexts.
Conclusions
Research on the HH pathway has uncovered unexpected principles in signal transduction: the covalent modification of ligands by cholesterol, the use of primary cilia to organize signaling, the regulation of a transcription factor by partial proteolysis and the use of cholesterol as a second messenger to regulate a cell-surface receptor. Several mechanistic questions remain to be fully answered, including the questions of how PTCH inhibits SMO activity and how SMO relays the signal to the GLI transcription factors. Further work is necessary to understand how the biochemical mechanisms of HH signaling are integrated with the cell biology of cilia trafficking and cilia localization. For example, why are cilia required for the formation of activator and repressor forms of the GLI proteins? The discoveries of signaling components that modify HH signaling strength in specific tissues are also likely to reveal new mechanistic and biological insights. Finally, the use of in vitro systems to reconstitute various aspects of signaling in both cells and across tissues, in combination with computational analyses, will be necessary to fully understand how the formation and interpretation of the HH signaling gradient can pattern the diverse metazoan body plans that are seen across evolution.
Acknowledgements
We apologize to the investigators whose work we were unable to cite owing to space constraints and the investigators working in the many important areas of HH signaling we were unable to cover. We thank Ganesh Pusapati for comments on the manuscript, and Steven R. Chan and Bertie Ansell for help with illustrations.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
R.R. is supported by the National Institutes of Health (GM118082 and GM106078), C.S. is supported by Cancer Research UK (C20724/A14414 and C20724/A26752) and the European Research Council (647278), and J.H.K. is supported by the American Heart Association (19POST34380734). Deposited in PMC for release after 12 months.
References
- Allen B. L., Song J. Y., Izzi L., Althaus I. W., Kang J.-S., Charron F., Krauss R. S. and McMahon A. P. (2011). Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC in SHH pathway function. Dev. Cell 20, 775-787. 10.1016/j.devcel.2011.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood S. X., Li M., Lee A., Tang J. Y. and Oro A. E. (2013). GLI activation by atypical protein kinase C ι/λ regulates the growth of basal cell carcinomas. Nature 494, 484-488. 10.1038/nature11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayers K. L. and Thérond P. P. (2010). Evaluating Smoothened as a G-protein-coupled receptor for Hedgehog signalling. Trends Cell Biol. 20, 287-298. 10.1016/j.tcb.2010.02.002 [DOI] [PubMed] [Google Scholar]
- Aza-Blanc P., Ramírez-Weber F.-A., Laget M. P., Schwartz C. and Kornberg T. B. (1997). Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell 89, 1043-1053. 10.1016/S0092-8674(00)80292-5 [DOI] [PubMed] [Google Scholar]
- Bangs F. and Anderson K. V. (2017). Primary cilia and mammalian hedgehog signaling. Cold Spring Harb. Perspect. Biol. 9, a028175 10.1101/cshperspect.a028175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzi M., Berenguer J., Menendez A., Alvarez-Rodriguez R. and Pons S. (2010). Sonic-hedgehog-mediated proliferation requires the localization of PKA to the cilium base. J. Cell Sci. 123, 62-69. 10.1242/jcs.060020 [DOI] [PubMed] [Google Scholar]
- Bazan J. F. and de Sauvage F. J. (2009). Structural ties between cholesterol transport and morphogen signaling. Cell 138, 1055-1056. 10.1016/j.cell.2009.09.006 [DOI] [PubMed] [Google Scholar]
- Belgacem Y. H. and Borodinsky L. N. (2011). Sonic hedgehog signaling is decoded by calcium spike activity in the developing spinal cord. Proc. Natl. Acad. Sci. USA 108, 4482-4487. 10.1073/pnas.1018217108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidet M., Joubert O., Lacombe B., Ciantar M., Nehmé R., Mollat P., Brétillon L., Faure H., Bittman R., Ruat M. et al. (2011). The hedgehog receptor patched is involved in cholesterol transport. PLoS ONE 6, e23834 10.1371/journal.pone.0023834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff M., Gradilla A.-C., Seijo I., Andrés G., Rodríguez-Navas C., González-Méndez L. and Guerrero I. (2013). Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nat. Cell Biol. 15, 1269-1281. 10.1038/ncb2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop B., Aricescu A. R., Harlos K., O'Callaghan C. A., Jones E. Y. and Siebold C. (2009). Structural insights into hedgehog ligand sequestration by the human hedgehog-interacting protein HHIP. Nat. Struct. Mol. Biol. 16, 698-703. 10.1038/nsmb.1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blassberg R., Macrae J. I., Briscoe J. and Jacob J. (2016). Reduced cholesterol levels impair Smoothened activation in Smith-Lemli-Opitz syndrome. Hum. Mol. Genet. 25, 693-705. 10.1093/hmg/ddv507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosanac I., Maun H. R., Scales S. J., Wen X., Lingel A., Bazan J. F., de Sauvage F. J., Hymowitz S. G. and Lazarus R. A. (2009). The structure of SHH in complex with HHIP reveals a recognition role for the Shh pseudo active site in signaling. Nat. Struct. Mol. Biol. 16, 691-697. 10.1038/nsmb.1632 [DOI] [PubMed] [Google Scholar]
- Breslow D. K., Hoogendoorn S., Kopp A. R., Morgens D. W., Vu B. K., Kennedy M. C., Han K., Li A., Hess G. T., Bassik M. C. et al. (2018). A CRISPR-based screen for Hedgehog signaling provides insights into ciliary function and ciliopathies. Nat. Genet. 50, 460-471. 10.1038/s41588-018-0054-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J. and Therond P. P. (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14, 416-429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Briscoe J., Chen Y., Jessell T. M. and Struhl G. (2001). A hedgehog-insensitive form of patched provides evidence for direct long-range morphogen activity of sonic hedgehog in the neural tube. Mol. Cell 7, 1279-1291. 10.1016/S1097-2765(01)00271-4 [DOI] [PubMed] [Google Scholar]
- Buglino J. A. and Resh M. D. (2008). Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J. Biol. Chem. 283, 22076-22088. 10.1074/jbc.M803901200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke R., Nellen D., Bellotto M., Hafen E., Senti K. A., Dickson B. J. and Basler K. (1999). Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell 99, 803-815. 10.1016/S0092-8674(00)81677-3 [DOI] [PubMed] [Google Scholar]
- Byrne E. F., Sircar R., Miller P. S., Hedger G., Luchetti G., Nachtergaele S., Tully M. D., Mydock-McGrane L., Covey D. F., Rambo R. P. et al. (2016). Structural basis of Smoothened regulation by its extracellular domains. Nature 535, 517-522. 10.1038/nature18934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne E. F., Luchetti G., Rohatgi R. and Siebold C. (2018). Multiple ligand binding sites regulate the Hedgehog signal transducer Smoothened in vertebrates. Curr. Opin. Cell Biol. 51, 81-88. 10.1016/j.ceb.2017.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter D., Stone D. M., Brush J., Ryan A., Armanini M., Frantz G., Rosenthal A. and de Sauvage F. J. (1998). Characterization of two patched receptors for the vertebrate hedgehog protein family. Proc. Natl. Acad. Sci. USA 95, 13630-13634. 10.1073/pnas.95.23.13630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstea E. D., Morris J. A., Coleman K. G., Loftus S. K., Zhang D., Cummings C., Gu J., Rosenfeld M. A., Pavan W. J., Krizman D. B. et al. (1997). Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science 277, 228-231. 10.1126/science.277.5323.228 [DOI] [PubMed] [Google Scholar]
- Caspary T., García-García M. J., Huangfu D., Eggenschwiler J. T., Wyler M. R., Rakeman A. S., Alcorn H. L. and Anderson K. V. (2002). Mouse Dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Curr. Biol. 12, 1628-1632. 10.1016/S0960-9822(02)01147-8 [DOI] [PubMed] [Google Scholar]
- Chamoun Z., Mann R. K., Nellen D., von Kessler D. P., Bellotto M., Beachy P. A. and Basler K. (2001). Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science 293, 2080-2084. 10.1126/science.1064437 [DOI] [PubMed] [Google Scholar]
- Charron F., Stein E., Jeong J., McMahon A. P. and Tessier-Lavigne M. (2003). The morphogen sonic hedgehog is an axonal chemoattractant that collaborates with netrin-1 in midline axon guidance. Cell 113, 11-23. 10.1016/S0092-8674(03)00199-5 [DOI] [PubMed] [Google Scholar]
- Chen Y. and Struhl G. (1996). Dual roles for patched in sequestering and transducing Hedgehog. Cell 87, 553-563. 10.1016/S0092-8674(00)81374-4 [DOI] [PubMed] [Google Scholar]
- Chen J. K., Taipale J., Cooper M. K. and Beachy P. A. (2002a). Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743-2748. 10.1101/gad.1025302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. K., Taipale J., Young K. E., Maiti T. and Beachy P. A. (2002b). Small molecule modulation of Smoothened activity. Proc. Natl. Acad. Sci. USA 99, 14071-14076. 10.1073/pnas.182542899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.-H., Li Y.-J., Kawakami T., Xu S.-M. and Chuang P.-T. (2004). Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes Dev. 18, 641-659. 10.1101/gad.1185804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.-H., Wilson C. W., Li Y.-J., Law K. K. L., Lu C.-S., Gacayan R., Zhang X., Hui C.-C. and Chuang P.-T. (2009). Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 23, 1910-1928. 10.1101/gad.1794109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Li S., Tong C., Zhao Y., Wang B., Liu Y., Jia J. and Jiang J. (2010). G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes Dev. 24, 2054-2067. 10.1101/gad.1948710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Sasai N., Ma G., Yue T., Jia J., Briscoe J. and Jiang J. (2011). Sonic Hedgehog dependent phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 9, e1001083 10.1371/journal.pbio.1001083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Huang H., Hatori R. and Kornberg T. B. (2017). Essential basal cytonemes take up Hedgehog in the Drosophila wing imaginal disc. Development 144, 3134-3144. 10.1242/dev.149856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla P., Xiao L., Kazanietz M. G. and Riobo N. A. (2010). Hedgehog proteins activate pro-angiogenic responses in endothelial cells through non-canonical signaling pathways. Cell Cycle 9, 570-579. 10.4161/cc.9.3.10591 [DOI] [PubMed] [Google Scholar]
- Chuang P. T. and McMahon A. P. (1999). Vertebrate Hedgehog signalling modulated by induction of a Hedgehog-binding protein. Nature 397, 617-621. 10.1038/17611 [DOI] [PubMed] [Google Scholar]
- Cooper M. K., Porter J. A., Young K. E. and Beachy P. A. (1998). Teratogen-mediated inhibition of target tissue response to Shh signaling. Science 280, 1603-1607. 10.1126/science.280.5369.1603 [DOI] [PubMed] [Google Scholar]
- Cooper M. K., Wassif C. A., Krakowiak P. A., Taipale J., Gong R., Kelley R. I., Porter F. D. and Beachy P. A. (2003). A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat. Genet. 33, 508-513. 10.1038/ng1134 [DOI] [PubMed] [Google Scholar]
- Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y. and Reiter J. F. (2005). Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018-1021. 10.1038/nature04117 [DOI] [PubMed] [Google Scholar]
- Corcoran R. B. and Scott M. P. (2006). Oxysterols stimulate Sonic hedgehog signal transduction and proliferation of medulloblastoma cells. Proc. Natl. Acad. Sci. USA 103, 8408-8413. 10.1073/pnas.0602852103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creanga A., Glenn T. D., Mann R. K., Saunders A. M., Talbot W. S. and Beachy P. A. (2012). Scube/You activity mediates release of dually lipid-modified Hedgehog signal in soluble form. Genes Dev. 26, 1312-1325. 10.1101/gad.191866.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. P. and Ioannou Y. A. (2000). Topological analysis of Niemann-Pick C1 protein reveals that the membrane orientation of the putative sterol-sensing domain is identical to those of 3-hydroxy-3-methylglutaryl-CoA reductase and sterol regulatory element binding protein cleavage-activating protein. J. Biol. Chem. 275, 24367-24374. 10.1074/jbc.M002184200 [DOI] [PubMed] [Google Scholar]
- DeCamp D. L., Thompson T. M., de Sauvage F. J. and Lerner M. R. (2000). Smoothened activates Galphai-mediated signaling in frog melanophores. J. Biol. Chem. 275, 26322-26327. 10.1074/jbc.M004055200 [DOI] [PubMed] [Google Scholar]
- Denef N., Neubüser D., Perez L. and Cohen S. M. (2000). Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell 102, 521-531. 10.1016/S0092-8674(00)00056-8 [DOI] [PubMed] [Google Scholar]
- Dessaud E., Yang L. L., Hill K., Cox B., Ulloa F., Ribeiro A., Mynett A., Novitch B. G. and Briscoe J. (2007). Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature 450, 717-720. 10.1038/nature06347 [DOI] [PubMed] [Google Scholar]
- Dijkgraaf G. J., Alicke B., Weinmann L., Januario T., West K., Modrusan Z., Burdick D., Goldsmith R., Robarge K., Sutherlin D. et al. (2011). Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 71, 435-444. 10.1158/0008-5472.CAN-10-2876 [DOI] [PubMed] [Google Scholar]
- Dorn K. V., Hughes C. E. and Rohatgi R. (2012). A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev. Cell 23, 823-835. 10.1016/j.devcel.2012.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer J. R., Sever N., Carlson M., Nelson S. F., Beachy P. A. and Parhami F. (2007). Oxysterols are novel activators of the hedgehog signaling pathway in pluripotent mesenchymal cells. J. Biol. Chem. 282, 8959-8968. 10.1074/jbc.M611741200 [DOI] [PubMed] [Google Scholar]
- Eccles R. L., Czajkowski M. T., Barth C., Müller P. M., McShane E., Grunwald S., Beaudette P., Mecklenburg N., Volkmer R., Zühlke K. et al. (2016). Bimodal antagonism of PKA signalling by ARHGAP36. Nat. Commun. 7, 12963 10.1038/ncomms12963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echelard Y., Epstein D. J., St-Jacques B., Shen L., Mohler J., McMahon J. A. and McMahon A. P. (1993). Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75, 1417-1430. 10.1016/0092-8674(93)90627-3 [DOI] [PubMed] [Google Scholar]
- Fan C. M., Porter J. A., Chiang C., Chang D. T., Beachy P. A. and Tessier-Lavigne M. (1995). Long-range sclerotome induction by sonic hedgehog: direct role of the amino-terminal cleavage product and modulation by the cyclic AMP signaling pathway. Cell 81, 457-465. 10.1016/0092-8674(95)90398-4 [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetsky M., Zhang X. M., Bottega S., Guicherit O., Wichterle H., Dudek H., Bumcrot D., Wang F. Y., Jones S., Shulok J. et al. (2002). Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J. Biol. 1, 10 10.1186/1475-4924-1-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Gonzalo F. R., Corbit K. C., Sirerol-Piquer M. S., Ramaswami G., Otto E. A., Noriega T. R., Seol A. D., Robinson J. F., Bennett C. L., Josifova D. J. et al. (2011). A transition zone complex regulates mammalian ciliogenesis and ciliary membrane composition. Nat. Genet. 43, 776-784. 10.1038/ng.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Milenkovic L., Suyama K., Hartl T., Purzner T., Winans A., Meyer T. and Scott M. P. (2015). Phosphodiesterase 4D acts downstream of Neuropilin to control Hedgehog signal transduction and the growth of medulloblastoma. Elife 4, e07068 10.7554/eLife.07068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X., Qian H., Cao P., Zhao X., Zhou Q., Lei J. and Yan N. (2018). Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science 361, eaas8935 10.1126/science.aas8935 [DOI] [PubMed] [Google Scholar]
- Goodrich L. V., Johnson R. L., Milenkovic L., McMahon J. A. and Scott M. P. (1996). Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 10, 301-312. 10.1101/gad.10.3.301 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M. and McMahon A. P. (1998). The effect of pertussis toxin on zebrafish development: a possible role for inhibitory G-proteins in hedgehog signaling. Dev. Biol. 194, 166-171. 10.1006/dbio.1997.8796 [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M., Bitgood M. J. and McMahon A. P. (1996). Protein kinase A is a common negative regulator of Hedgehog signaling in the vertebrate embryo. Genes Dev. 10, 647-658. 10.1101/gad.10.6.647 [DOI] [PubMed] [Google Scholar]
- Hausmann G., von Mering C. and Basler K. (2009). The hedgehog signaling pathway: where did it come from? PLoS Biol. 7, e1000146 10.1371/journal.pbio.1000146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycraft C. J., Banizs B., Aydin-Son Y., Zhang Q., Michaud E. J. and Yoder B. K. (2005). Gli2 and Gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 1, e53 10.1371/journal.pgen.0010053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M., Subramanian R., Bangs F., Omelchenko T., Liem K. F. Jr, Kapoor T. M. and Anderson K. V. (2014a). The kinesin-4 protein KIF7 regulates mammalian hedgehog signaling by organizing the cilia tip compartment. Nat. Cell Biol. 16, 663-672. 10.1038/ncb2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Zhang L., Chen Y., Remke M., Shih D., Lu F., Wang H., Deng Y., Yu Y., Xia Y. et al. (2014b). The G protein α subunit Gαs is a tumor suppressor in Sonic hedgehog-driven medulloblastoma. Nat. Med. 20, 1035-1042. 10.1038/nm.3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedger G., Koldsø H., Chavent M., Siebold C., Rohatgi R. and Sansom M. S. P. (2019). Cholesterol interaction sites on the transmembrane domain of the hedgehog signal transducer and class F G protein-coupled receptor smoothened. Structure 27, 549-559. 10.1016/j.str.2018.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz A. M., Griffiths S. C., Davis S. J., Bishop B., Siebold C. and Allen B. L. (2015). Secreted HHIP1 interacts with heparan sulfate and regulates Hedgehog ligand localization and function. J. Cell Biol. 209, 739-757. 10.1083/jcb.201411024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Roelink H. and McKnight G. S. (2002). Protein kinase A deficiency causes axially localized neural tube defects in mice. J. Biol. Chem. 277, 19889-19896. 10.1074/jbc.M111412200 [DOI] [PubMed] [Google Scholar]
- Huang P., Nedelcu D., Watanabe M., Jao C., Kim Y., Liu J. and Salic A. (2016). Cellular cholesterol directly activates smoothened in hedgehog signaling. Cell 166, 1176-1187.e14. 10.1016/j.cell.2016.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Zheng S., Wierbowski B. M., Kim Y., Nedelcu D., Aravena L., Liu J., Kruse A. C. and Salic A. (2018). Structural basis of smoothened activation in hedgehog signaling. Cell 175, 295-297. 10.1016/j.cell.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huangfu D., Liu A., Rakeman A. S., Murcia N. S., Niswander L. and Anderson K. V. (2003). Hedgehog signalling in the mouse requires intraflagellar transport proteins. Nature 426, 83-87. 10.1038/nature02061 [DOI] [PubMed] [Google Scholar]
- Hui C. C. and Angers S. (2011). Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol. 27, 513-537. 10.1146/annurev-cellbio-092910-154048 [DOI] [PubMed] [Google Scholar]
- Hui C. C., Slusarski D., Platt K. A., Holmgren R. and Joyner A. L. (1994). Expression of three mouse homologs of the Drosophila segment polarity gene cubitus interruptus, Gli, Gli-2, and Gli-3, in ectoderm- and mesoderm-derived tissues suggests multiple roles during postimplantation development. Dev. Biol. 162, 402-413. 10.1006/dbio.1994.1097 [DOI] [PubMed] [Google Scholar]
- Humke E. W., Dorn K. V., Milenkovic L., Scott M. P. and Rohatgi R. (2010). The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 24, 670-682. 10.1101/gad.1902910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzicker E. G., Estay I. S., Zhen H., Lokteva L. A., Jackson P. K. and Oro A. E. (2006). Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 20, 276-281. 10.1101/gad.1380906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S.-H., White K. A., Somatilaka B. N., Shelton J. M., Richardson J. A. and Mukhopadhyay S. (2018). The G protein-coupled receptor Gpr161 regulates forelimb formation, limb patterning and skeletal morphogenesis in a primary cilium-dependent manner. Development 145, dev154054 10.1242/dev.154054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes M., Porter J. A., Chiang C., Chang D., Tessier-Lavigne M., Beachy P. A. and Rosenthal A. (1995). Induction of midbrain dopaminergic neurons by Sonic hedgehog. Neuron 15, 35-44. 10.1016/0896-6273(95)90062-4 [DOI] [PubMed] [Google Scholar]
- Iglesias-Bartolome R., Torres D., Marone R., Feng X., Martin D., Simaan M., Chen M., Weinstein L. S., Taylor S. S., Molinolo A. A. et al. (2015). Inactivation of a Gα(s)-PKA tumour suppressor pathway in skin stem cells initiates basal-cell carcinogenesis. Nat. Cell Biol. 17, 793-803. 10.1038/ncb3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona J. P. and Roelink H. (2000). The role of cholesterol in Shh signaling and teratogen-induced holoprosencephaly. Cell. Mol. Life Sci. 57, 1709-1719. 10.1007/PL00000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incardona J. P., Gruenberg J. and Roelink H. (2002). Sonic hedgehog induces the segregation of patched and smoothened in endosomes. Curr. Biol. 12, 983-995. 10.1016/S0960-9822(02)00895-3 [DOI] [PubMed] [Google Scholar]
- Ingham P. W. (2018). From Drosophila segmentation to human cancer therapy. Development 145, dev168898 10.1242/dev.168898 [DOI] [PubMed] [Google Scholar]
- Ingham P. W., Taylor A. M. and Nakano Y. (1991). Role of the Drosophila patched gene in positional signalling. Nature 353, 184-187. 10.1038/353184a0 [DOI] [PubMed] [Google Scholar]
- Ingham P. W., Nystedt S., Nakano Y., Brown W., Stark D., van den Heuvel M. and Taylor A. M. (2000). Patched represses the Hedgehog signalling pathway by promoting modification of the Smoothened protein. Curr. Biol. 10, 1315-1318. 10.1016/S0960-9822(00)00755-7 [DOI] [PubMed] [Google Scholar]
- Izzi L., Lévesque M., Morin S., Laniel D., Wilkes B. C., Mille F., Krauss R. S., McMahon A. P., Allen B. L. and Charron F. (2011). Boc and Gas1 each form distinct Shh receptor complexes with Ptch1 and are required for Shh-mediated cell proliferation. Dev. Cell 20, 788-801. 10.1016/j.devcel.2011.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda C. Y., Waghray D., Levin A. M., Thomas C. and Garcia K. C. (2012). Structural basis of Wnt recognition by Frizzled. Science 337, 59-64. 10.1126/science.1222879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavran J. M., Ward M. D., Oladosu O. O., Mulepati S. and Leahy D. J. (2010). All mammalian Hedgehog proteins interact with cell adhesion molecule, down-regulated by oncogenes (CDO) and brother of CDO (BOC) in a conserved manner. J. Biol. Chem. 285, 24584-24590. 10.1074/jbc.M110.131680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T., Kawcak T., Li Y.-J., Zhang W., Hu Y. and Chuang P.-T. (2002). Mouse dispatched mutants fail to distribute hedgehog proteins and are defective in hedgehog signaling. Development 129, 5753-5765. 10.1242/dev.00178 [DOI] [PubMed] [Google Scholar]
- Kim J., Kato M. and Beachy P. A. (2009). Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. USA 106, 21666-21671. 10.1073/pnas.0912180106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss S., Concordet J.-P. and Ingham P. W. (1993). A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75, 1431-1444. 10.1016/0092-8674(93)90628-4 [DOI] [PubMed] [Google Scholar]
- Kumar N., Su C.-C., Chou T.-H., Radhakrishnan A., Delmar J. A., Rajashankar K. R. and Yu E. W. (2017). Crystal structures of the Burkholderia multivorans hopanoid transporter HpnN. Proc. Natl. Acad. Sci. USA 114, 6557-6562. 10.1073/pnas.1619660114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. J., Ekker S. C., von Kessler D. P., Porter J. A., Sun B. I. and Beachy P. A. (1994). Autoproteolysis in hedgehog protein biogenesis. Science 266, 1528-1537. 10.1126/science.7985023 [DOI] [PubMed] [Google Scholar]
- Lee R. T. H., Zhao Z. and Ingham P. W. (2016). Hedgehog signalling. Development 143, 367-372. 10.1242/dev.120154 [DOI] [PubMed] [Google Scholar]
- Lewis P. M., Dunn M. P., McMahon J. A., Logan M., Martin J. F., St-Jacques B. and McMahon A. P. (2001). Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell 105, 599-612. 10.1016/S0092-8674(01)00369-5 [DOI] [PubMed] [Google Scholar]
- Li P., Markson J. S., Wang S., Chen S., Vachharajani V. and Elowitz M. B. (2018). Morphogen gradient reconstitution reveals Hedgehog pathway design principles. Science 360, 543-548. 10.1126/science.aao0645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litingtung Y. and Chiang C. (2000). Specification of ventral neuron types is mediated by an antagonistic interaction between Shh and Gli3. Nat. Neurosci. 3, 979-985. 10.1038/79916 [DOI] [PubMed] [Google Scholar]
- Loftus S. K., Morris J. A., Carstea E. D., Gu J. Z., Cummings C., Brown A., Ellison J., Ohno K., Rosenfeld M. A., Tagle D. A. et al. (1997). Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science 277, 232-235. 10.1126/science.277.5323.232 [DOI] [PubMed] [Google Scholar]
- Low W.-C., Wang C., Pan Y., Huang X.-Y., Chen J. K. and Wang B. (2008). The decoupling of Smoothened from Gαi proteins has little effect on Gli3 protein processing and Hedgehog-regulated chick neural tube patterning. Dev. Biol. 321, 188-196. 10.1016/j.ydbio.2008.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Liu S. and Kornberg T. B. (2006). The C-terminal tail of the Hedgehog receptor Patched regulates both localization and turnover. Genes Dev. 20, 2539-2551. 10.1101/gad.1461306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchetti G., Sircar R., Kong J. H., Nachtergaele S., Sagner A., Byrne E. F., Covey D. F., Siebold C. and Rohatgi R. (2016). Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. Elife 5, e20304 10.7554/eLife.20304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Erkner A., Gong R., Yao S., Taipale J., Basler K. and Beachy P. A. (2002). Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell 111, 63-75. 10.1016/S0092-8674(02)00977-7 [DOI] [PubMed] [Google Scholar]
- Manikowski D., Kastl P. and Grobe K. (2018). Taking the Occam's razor approach to hedgehog lipidation and its role in development. J. Dev. Biol. 6, E3 10.3390/jdb6010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigo V., Davey R. A., Zuo Y., Cunningham J. M. and Tabin C. J. (1996a). Biochemical evidence that patched is the Hedgehog receptor. Nature 384, 176-179. 10.1038/384176a0 [DOI] [PubMed] [Google Scholar]
- Marigo V., Scott M. P., Johnson R. L., Goodrich L. V. and Tabin C. J. (1996b). Conservation in hedgehog signaling: induction of a chicken patched homolog by Sonic hedgehog in the developing limb. Development 122, 1225-1233. [DOI] [PubMed] [Google Scholar]
- Matise M. P., Epstein D. J., Park H. L., Platt K. A. and Joyner A. L. (1998). Gli2 is required for induction of floor plate and adjacent cells, but not most ventral neurons in the mouse central nervous system. Development 125, 2759-2770. [DOI] [PubMed] [Google Scholar]
- McLellan J. S., Zheng X., Hauk G., Ghirlando R., Beachy P. A. and Leahy D. J. (2008). The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla. Nature 455, 979-983. 10.1038/nature07358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon A. P., Ingham P. W. and Tabin C. J. (2003). Developmental roles and clinical significance of hedgehog signaling. Curr. Top. Dev. Biol. 53, 1-114. 10.1016/S0070-2153(03)53002-2 [DOI] [PubMed] [Google Scholar]
- Meloni A. R., Fralish G. B., Kelly P., Salahpour A., Chen J. K., Wechsler-Reya R. J., Lefkowitz R. J. and Caron M. G. (2006). Smoothened signal transduction is promoted by G protein-coupled receptor kinase 2. Mol. Cell. Biol. 26, 7550-7560. 10.1128/MCB.00546-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methot N. and Basler K. (1999). Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell 96, 819-831. 10.1016/S0092-8674(00)80592-9 [DOI] [PubMed] [Google Scholar]
- Mick D. U., Rodrigues R. B., Leib R. D., Adams C. M., Chien A. S., Gygi S. P. and Nachury M. V. (2015). Proteomics of primary cilia by proximity labeling. Dev. Cell 35, 497-512. 10.1016/j.devcel.2015.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza A. N., McKellar S. A., Urman N. M., Brown A. S., Hollmig T., Aasi S. Z. and Oro A. E. (2019). LAP2 proteins chaperone GLI1 movement between the lamina and chromatin to regulate transcription. Cell 176, 198-212.e15. 10.1016/j.cell.2018.10.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B. S., Stepanchick A. N., Tewson P. H., Hartle C. M., Zhang J., Quinn A. M., Hughes T. E. and Mirshahi T. (2016). Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc. Natl. Acad. Sci. USA 113, 13069-13074. 10.1073/pnas.1602393113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motoyama J., Takabatake T., Takeshima K. and Hui C. (1998). Ptch2, a second mouse Patched gene is co-expressed with Sonic hedgehog. Nat. Genet. 18, 104-106. 10.1038/ng0298-104 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S., Wen X., Ratti N., Loktev A., Rangell L., Scales S. J. and Jackson P. K. (2013). The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell 152, 210-223. 10.1016/j.cell.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Myers B. R., Sever N., Chong Y. C., Kim J., Belani J. D., Rychnovsky S., Bazan J. F. and Beachy P. A. (2013). Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev. Cell 26, 346-357. 10.1016/j.devcel.2013.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers B. R., Neahring L., Zhang Y., Roberts K. J. and Beachy P. A. (2017). Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc. Natl. Acad. Sci. USA 114, E11141-E11150. 10.1073/pnas.1717891115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S., Mydock L. K., Krishnan K., Rammohan J., Schlesinger P. H., Covey D. F. and Rohatgi R. (2012). Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat. Chem. Biol. 8, 211-220. 10.1038/nchembio.765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachtergaele S., Whalen D. M., Mydock L. K., Zhao Z., Malinauskas T., Krishnan K., Ingham P. W., Covey D. F., Siebold C. and Rohatgi R. (2013). Structure and function of the Smoothened extracellular domain in vertebrate Hedgehog signaling. Elife 2, e01340 10.7554/eLife.01340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelcu D., Liu J., Xu Y., Jao C. and Salic A. (2013). Oxysterol binding to the extracellular domain of Smoothened in Hedgehog signaling. Nat. Chem. Biol. 9, 557-564. 10.1038/nchembio.1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis E. and Hui C.-C. (2005). Hedgehog signaling and congenital malformations. Clin. Genet. 67, 193-208. 10.1111/j.1399-0004.2004.00360.x [DOI] [PubMed] [Google Scholar]
- Nieuwenhuis E., Motoyama J., Barnfield P. C., Yoshikawa Y., Zhang X., Mo R., Crackower M. A. and Hui C.-C. (2006). Mice with a targeted mutation of patched2 are viable but develop alopecia and epidermal hyperplasia. Mol. Cell. Biol. 26, 6609-6622. 10.1128/MCB.00295-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P., Zhujiang A., Youssef M. and Waschek J. A. (2013). Interaction of PACAP with Sonic hedgehog reveals complex regulation of the hedgehog pathway by PKA. Cell. Signal. 25, 2222-2230. 10.1016/j.cellsig.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewiadomski P., Kong J. H., Ahrends R., Ma Y., Humke E. W., Khan S., Teruel M. N., Novitch B. G. and Rohatgi R. (2014). Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell Rep. 6, 168-181. 10.1016/j.celrep.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C. and Wieschaus E. (1980). Mutations affecting segment number and polarity in Drosophila. Nature 287, 795-801. 10.1038/287795a0 [DOI] [PubMed] [Google Scholar]
- Ocbina P. J. and Anderson K. V. (2008). Intraflagellar transport, cilia, and mammalian Hedgehog signaling: analysis in mouse embryonic fibroblasts. Dev. Dyn. 237, 2030-2038. 10.1002/dvdy.21551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal K., Hwang S.-H., Somatilaka B., Badgandi H., Jackson P. K., DeFea K. and Mukhopadhyay S. (2016). Smoothened determines β-arrestin--mediated removal of the G protein--coupled receptor Gpr161 from the primary cilium. J. Cell Biol. 212, 861-875. 10.1083/jcb.201506132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepinsky R. B., Zeng C., Wen D., Rayhorn P., Baker D. P., Williams K. P., Bixler S. A., Ambrose C. M., Garber E. A., Miatkowski K. et al. (1998). Identification of a palmitic acid-modified form of human Sonic hedgehog. J. Biol. Chem. 273, 14037-14045. 10.1074/jbc.273.22.14037 [DOI] [PubMed] [Google Scholar]
- Petrov K., Wierbowski B. M. and Salic A. (2017). Sending and receiving hedgehog signals. Annu. Rev. Cell Dev. Biol. 33, 145-168. 10.1146/annurev-cellbio-100616-060847 [DOI] [PubMed] [Google Scholar]
- Pfeffer S. R. (2019). NPC intracellular cholesterol transporter 1 (NPC1)-mediated cholesterol export from lysosomes. J. Biol. Chem. 293, 1706-1709. 10.1074/jbc.TM118.004165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipp M., Fralish G. B., Meloni A. R., Chen W., MacInnes A. W., Barak L. S. and Caron M. G. (2008). Smoothened signaling in vertebrates is facilitated by a G protein-coupled receptor kinase. Mol. Biol. Cell 19, 5478-5489. 10.1091/mbc.e08-05-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J. A., Young K. E. and Beachy P. A. (1996). Cholesterol modification of hedgehog signaling proteins in animal development. Science 274, 255-259. 10.1126/science.274.5285.255 [DOI] [PubMed] [Google Scholar]
- Pusapati G. V., Hughes C. E., Dorn K. V., Zhang D., Sugianto P., Aravind L. and Rohatgi R. (2014). EFCAB7 and IQCE regulate hedgehog signaling by tethering the EVC-EVC2 complex to the base of primary cilia. Dev. Cell 28, 483-496. 10.1016/j.devcel.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusapati G. V., Kong J. H., Patel B. B., Krishnan A., Sagner A., Kinnebrew M., Briscoe J., Aravind L. and Rohatgi R. (2018a). CRISPR screens uncover genes that regulate target cell sensitivity to the morphogen sonic hedgehog. Dev. Cell 44, 271 10.1016/j.devcel.2018.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusapati G. V., Kong J. H., Patel B. B., Gouti M., Sagner A., Sircar R., Luchetti G., Ingham P. W., Briscoe J. and Rohatgi R. (2018b). G protein-coupled receptors control the sensitivity of cells to the morphogen Sonic Hedgehog. Sci. Signal. 11 10.1126/scisignal.aao5749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Schmiege P., Coutavas E., Wang J. and Li X. (2018a). Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature 560, 128-132. 10.1038/s41586-018-0308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X., Schmiege P., Coutavas E. and Li X. (2018b). Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science 362 10.1126/science.aas8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H., Cao P., Hu M., Gao S., Yan N. and Gong X. (2018). Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. bioRxiv 461491 10.1101/461491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rack P. G., Ni J., Payumo A. Y., Nguyen V., Crapster J. A., Hovestadt V., Kool M., Jones D. T. W., Mich J. K., Firestone A. J. et al. (2014). Arhgap36-dependent activation of Gli transcription factors. Proc. Natl. Acad. Sci. USA 111, 11061-11066. 10.1073/pnas.1322362111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh D. R. and Reiter J. F. (2019). Misactivation of Hedgehog signaling causes inherited and sporadic cancers. J. Clin. Invest. 129, 465-475. 10.1172/JCI120850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raleigh D. R., Sever N., Choksi P. K., Sigg M. A., Hines K. M., Thompson B. M., Elnatan D., Jaishankar P., Bisignano P., Garcia-Gonzalo F. R. et al. (2018). Cilia-associated oxysterols activate smoothened. Mol. Cell 72, 316-327.e5. 10.1016/j.molcel.2018.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Weber F.-A. and Kornberg T. B. (1999). Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell 97, 599-607. 10.1016/S0092-8674(00)80771-0 [DOI] [PubMed] [Google Scholar]
- Rana R., Carroll C. E., Lee H. J., Bao J., Marada S., Grace C. R., Guibao C. D., Ogden S. K. and Zheng J. J. (2013). Structural insights into the role of the Smoothened cysteine-rich domain in Hedgehog signalling. Nat. Commun. 4, 2965 10.1038/ncomms3965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranieri N., Ruel L., Gallet A., Raisin S. and Thérond P. P. (2012). Distinct phosphorylations on kinesin costal-2 mediate differential hedgehog signaling strength. Dev. Cell 22, 279-294. 10.1016/j.devcel.2011.12.002 [DOI] [PubMed] [Google Scholar]
- Regard J. B., Malhotra D., Gvozdenovic-Jeremic J., Josey M., Chen M., Weinstein L. S., Lu J., Shore E. M., Kaplan F. S. and Yang Y. (2013). Activation of Hedgehog signaling by loss of GNAS causes heterotopic ossification. Nat. Med. 19, 1505-1512. 10.1038/nm.3314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle R. D., Johnson R. L., Laufer E. and Tabin C. (1993). Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75, 1401-1416. 10.1016/0092-8674(93)90626-2 [DOI] [PubMed] [Google Scholar]
- Riobo N. A., Saucy B., Dilizio C. and Manning D. R. (2006). Activation of heterotrimeric G proteins by Smoothened. Proc. Natl. Acad. Sci. USA 103, 12607-12612. 10.1073/pnas.0600880103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roelink H., Augsburger A., Heemskerk J., Korzh V., Norlin S., Ruiz i Altaba A., Tanabe Y., Placzek M., Edlund T. and Jessell T. M. (1994). Floor plate and motor neuron induction by vhh-1, a vertebrate homolog of hedgehog expressed by the notochord. Cell 76, 761-775. 10.1016/0092-8674(94)90514-2 [DOI] [PubMed] [Google Scholar]
- Rohatgi R. and Snell W. J. (2010). The ciliary membrane. Curr. Opin. Cell Biol. 22, 541-546. 10.1016/j.ceb.2010.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L. and Scott M. P. (2007). Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372-376. 10.1126/science.1139740 [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Milenkovic L., Corcoran R. B. and Scott M. P. (2009). Hedgehog signal transduction by Smoothened: pharmacologic evidence for a 2-step activation process. Proc. Natl. Acad. Sci. USA 106, 3196-3201. 10.1073/pnas.0813373106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders T. A., Llagostera E. and Barna M. (2013). Specialized filopodia direct long-range transport of SHH during vertebrate tissue patterning. Nature 497, 628-632. 10.1038/nature12157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader E. K., Harstad K. G., Holmgren R. A. and Matouschek A. (2011). A three-part signal governs differential processing of Gli1 and Gli3 proteins by the proteasome. J. Biol. Chem. 286, 39051-39058. 10.1074/jbc.M111.274993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe H. J., Wang W., Hannoush R. N. and de Sauvage F. J. (2015). Regulation of the oncoprotein Smoothened by small molecules. Nat. Chem. Biol. 11, 246-255. 10.1038/nchembio.1776 [DOI] [PubMed] [Google Scholar]
- Shen F., Cheng L., Douglas A. E., Riobo N. A. and Manning D. R. (2013). Smoothened is a fully competent activator of the heterotrimeric G protein Gi. Mol. Pharmacol. 83, 691-697. 10.1124/mol.112.082511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada I. S., Hwang S.-H., Somatilaka B. N., Wang X., Skowron P., Kim J., Kim M., Shelton J. M., Rajaram V., Xuan Z. et al. (2018). Basal suppression of the sonic hedgehog pathway by the G-protein-coupled receptor Gpr161 restricts medulloblastoma pathogenesis. Cell Rep. 22, 1169-1184. 10.1016/j.celrep.2018.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamataki D., Ulloa F., Tsoni S. V., Mynett A. and Briscoe J. (2005). A gradient of Gli activity mediates graded Sonic Hedgehog signaling in the neural tube. Genes Dev. 19, 626-641. 10.1101/gad.325905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steck T. L. and Lange Y. (2018). Transverse distribution of plasma membrane bilayer cholesterol: picking sides. Traffic 19, 750-760. 10.1111/tra.12586 [DOI] [PubMed] [Google Scholar]
- Stone D. M., Hynes M., Armanini M., Swanson T. A., Gu Q., Johnson R. L., Scott M. P., Pennica D., Goddard A., Phillips H. et al. (1996). The tumour-suppressor gene patched encodes a candidate receptor for Sonic hedgehog. Nature 384, 129-134. 10.1038/384129a0 [DOI] [PubMed] [Google Scholar]
- Strigini M. and Cohen S. M. (1997). A Hedgehog activity gradient contributes to AP axial patterning of the Drosophila wing. Development 124, 4697-4705. [DOI] [PubMed] [Google Scholar]
- Su Y., Ospina J. K., Zhang J., Michelson A. P., Schoen A. M. and Zhu A. J. (2011). Sequential phosphorylation of smoothened transduces graded hedgehog signaling. Sci. Signal. 4, ra43 10.1126/scisignal.4159ec43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale J., Chen J. K., Cooper M. K., Wang B., Mann R. K., Milenkovic L., Scott M. P. and Beachy P. A. (2000). Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature 406, 1005-1009. 10.1038/35023008 [DOI] [PubMed] [Google Scholar]
- Taipale J., Cooper M. K., Maiti T. and Beachy P. A. (2002). Patched acts catalytically to suppress the activity of Smoothened. Nature 418, 892-897. 10.1038/nature00989 [DOI] [PubMed] [Google Scholar]
- Taylor F. R., Wen D., Garber E. A., Carmillo A. N., Baker D. P., Arduini R. M., Williams K. P., Weinreb P. H., Rayhorn P., Hronowski X. et al. (2001). Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry 40, 4359-4371. 10.1021/bi002487u [DOI] [PubMed] [Google Scholar]
- Tempe D., Casas M., Karaz S., Blanchet-Tournier M. F. and Concordet J. P. (2006). Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP. Mol. Cell. Biol. 26, 4316-4326. 10.1128/MCB.02183-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperino R., Amann S., Bayer M., McGee S. L., Loipetzberger A., Connor T., Jaeger C., Kammerer B., Winter L., Wiche G. et al. (2012). Hedgehog partial agonism drives Warburg-like metabolism in muscle and brown fat. Cell 151, 414-426. 10.1016/j.cell.2012.09.021 [DOI] [PubMed] [Google Scholar]
- Thomson M. and Gunawardena J. (2009). Unlimited multistability in multisite phosphorylation systems. Nature 460, 274-277. 10.1038/nature08102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng T. T., Gratwick K. S., Kollman J., Park D., Nies D. H., Goffeau A. and Saier M. H. Jr (1999). The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1, 107-125. [PubMed] [Google Scholar]
- Tukachinsky H., Lopez L. V. and Salic A. (2010). A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J. Cell Biol. 191, 415-428. 10.1083/jcb.201004108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H., Kuzmickas R. P., Jao C. Y., Liu J. and Salic A. (2012). Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2, 308-320. 10.1016/j.celrep.2012.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukachinsky H., Petrov K., Watanabe M. and Salic A. (2016). Mechanism of inhibition of the tumor suppressor Patched by Sonic Hedgehog. Proc. Natl. Acad. Sci. USA 113, E5866-E5875. 10.1073/pnas.1606719113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuson M., He M. and Anderson K. V. (2011). Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development 138, 4921-4930. 10.1242/dev.070805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twigg S. R., Lloyd D., Jenkins D., Elcioglu N. E., Cooper C. D., Al-Sannaa N., Annagur A., Gillessen-Kaesbach G., Huning I., Knight S. J. et al. (2012). Mutations in multidomain protein MEGF8 identify a Carpenter syndrome subtype associated with defective lateralization. Am. J. Hum. Genet. 91, 897-905. 10.1016/j.ajhg.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M., Li S.-P. and Taipale J. (2006). Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev. Cell 10, 177-186. 10.1016/j.devcel.2005.12.014 [DOI] [PubMed] [Google Scholar]
- Vuolo L., Herrera A., Torroba B., Menendez A. and Pons S. (2015). Ciliary adenylyl cyclases control the Hedgehog pathway. J. Cell Sci. 128, 2928-2937. 10.1242/jcs.172635 [DOI] [PubMed] [Google Scholar]
- Wang B. and Li Y. (2006). Evidence for the direct involvement of {beta}TrCP in Gli3 protein processing. Proc. Natl. Acad. Sci. USA 103, 33-38. 10.1073/pnas.0509927103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Fallon J. F. and Beachy P. A. (2000). Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell 100, 423-434. 10.1016/S0092-8674(00)80678-9 [DOI] [PubMed] [Google Scholar]
- Wang C., Wu H., Katritch V., Han G. W., Huang X. P., Liu W., Siu F. Y., Roth B. L., Cherezov V. and Stevens R. C. (2013). Structure of the human smoothened receptor bound to an antitumour agent. Nature 497, 338-343. 10.1038/nature12167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wu H., Evron T., Vardy E., Han G. W., Huang X. P., Hufeisen S. J., Mangano T. J., Urban D. J., Katritch V. et al. (2014). Structural basis for Smoothened receptor modulation and chemoresistance to anticancer drugs. Nat. Commun. 5, 4355 10.1038/ncomms5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wechsler-Reya R. J. and Scott M. P. (1999). Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22, 103-114. 10.1016/S0896-6273(00)80682-0 [DOI] [PubMed] [Google Scholar]
- Whalen D. M., Malinauskas T., Gilbert R. J. C. and Siebold C. (2013). Structural insights into proteoglycan-shaped Hedgehog signaling. Proc. Natl. Acad. Sci. USA 110, 16420-16425. 10.1073/pnas.1310097110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. P., Rayhorn P., Chi-Rosso G., Garber E. A., Strauch K. L., Horan G. S., Reilly J. O., Baker D. P., Taylor F. R., Koteliansky V. et al. (1999). Functional antagonists of sonic hedgehog reveal the importance of the N terminus for activity. J. Cell Sci. 112, 4405-4414. [DOI] [PubMed] [Google Scholar]
- Williams C. H., Hempel J. E., Hao J., Frist A. Y., Williams M. M., Fleming J. T., Sulikowski G. A., Cooper M. K., Chiang C. and Hong C. C. (2015). An in vivo chemical genetic screen identifies phosphodiesterase 4 as a pharmacological target for hedgehog signaling inhibition. Cell Rep. 11, 43-50. 10.1016/j.celrep.2015.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. C., Kozielewicz P., Kowalski-Jahn M., Petersen J., Bowin C.-F., Slodkowicz G., Marti-Solano M., Rodríguez D., Hot B., Okashah N. et al. (2019). A conserved molecular switch in Class F receptors regulates receptor activation and pathway selection. Nat. Commun. 10, 667 10.1038/s41467-019-08630-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Tang J.-J., Peng C., Wang Y., Fu L., Qiu Z.-P., Xiong Y., Yang L.-F., Cui H.-W., He X.-L. et al. (2017). Cholesterol modification of smoothened is required for hedgehog signaling. Mol. Cell 66, 154-162.e10. 10.1016/j.molcel.2017.02.015 [DOI] [PubMed] [Google Scholar]
- Yam P. T., Langlois S. D., Morin S. and Charron F. (2009). Sonic hedgehog guides axons through a noncanonical, Src-family-kinase-dependent signaling pathway. Neuron 62, 349-362. 10.1016/j.neuron.2009.03.022 [DOI] [PubMed] [Google Scholar]
- Yao H. H.-C. and Capel B. (2002). Disruption of testis cords by cyclopamine or forskolin reveals independent cellular pathways in testis organogenesis. Dev. Biol. 246, 356-365. 10.1006/dbio.2002.0663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Alpert D., Francis R., Chatterjee B., Yu Q., Tansey T., Sabol S. L., Cui C., Bai Y., Koriabine M. et al. (2009). Massively parallel sequencing identifies the gene Megf8 with ENU-induced mutation causing heterotaxy. Proc. Natl. Acad. Sci. USA 106, 3219-3224. 10.1073/pnas.0813400106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhao F., Wu Y., Yang J., Han G. W., Zhao S., Ishchenko A., Ye L., Lin X., Ding K. et al. (2017). Crystal structure of a multi-domain human smoothened receptor in complex with a super stabilizing ligand. Nat. Commun. 8, 15383 10.1038/ncomms15383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Bulkley D. P., Xin Y., Roberts K. J., Asarnow D. E., Sharma A., Myers B. R., Cho W., Cheng Y. and Beachy P. A. (2018). Structural basis for cholesterol transport-like activity of the hedgehog receptor patched. Cell 175, 1352-1364. 10.1016/j.cell.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Lee R. T. H., Pusapati G. V., Iyu A., Rohatgi R. and Ingham P. W. (2016). An essential role for Grk2 in Hedgehog signalling downstream of Smoothened. EMBO Rep. 17, 739-752. 10.15252/embr.201541532 [DOI] [PMC free article] [PubMed] [Google Scholar]