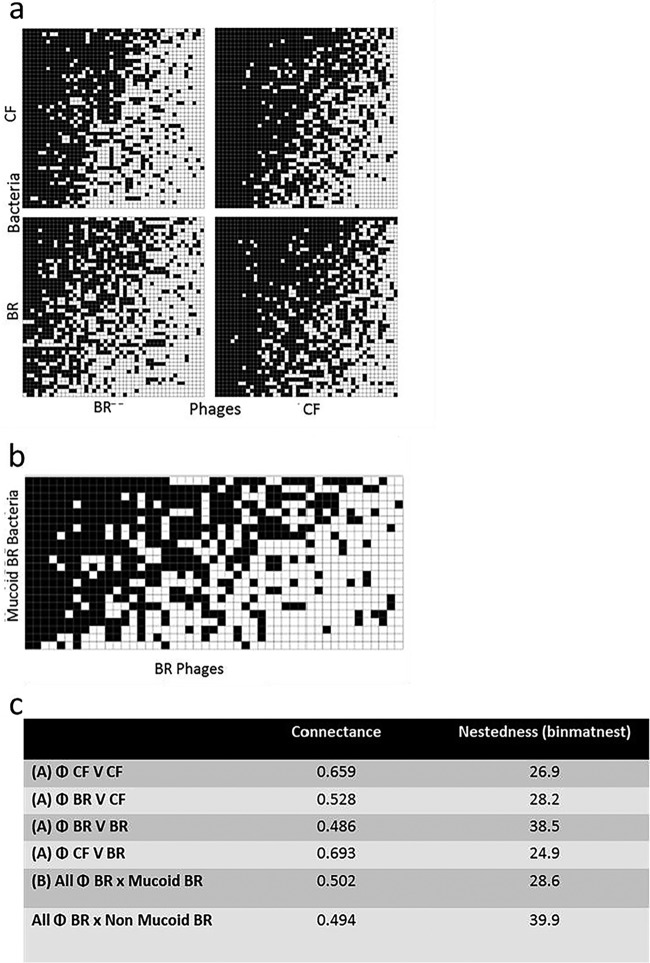
FIG 1.
Shows nestedness and connectance plots demonstrating that temperate phages from P. aeruginosa isolated from patients with advanced respiratory infection can infect a wider range of P. aeruginosa genotypes. The nestedness and connectance plots map the cross-infection data from mixed temperate phage communities induced from clinical P. aeruginosa isolates. (A) The binary nested network for the complete 94 phage lysates against 94 P. aeruginosa isolates, ordered by nestedness within equally sized quadrants (47 CF phages versus 47 CF P. aeruginosa isolates, 47 BR phages versus 47 CF P. aeruginosa isolates, 47 CF phages versus 47 BR P. aeruginosa isolates, and 47 BR phages versus 47 BR P. aeruginosa isolates). Each black square represents an individual interaction (infection event) between one phage strain (x axis) and one bacterial strain (y axis). White squares represent lack of infection. Note the CF-derived phages are capable of infecting BR P. aeruginosa isolates more frequently than BR phages can infect CF P. aeruginosa, illustrated through higher values for connectance (0.693). (B) The binary nested network representation of infection of the mucoid BR P. aeruginosa isolates (n = 22) with the entire cohort of mixed temperate phage communities induced from BR P. aeruginosa isolates (n = 47). (C) Nestedness and connectance values for the results shown in panels A and B.