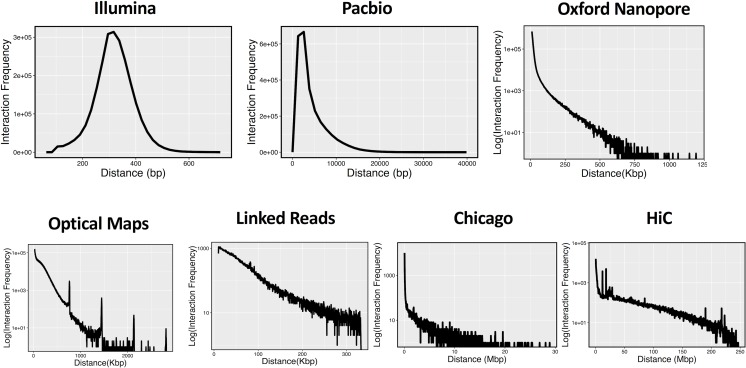
Fig 2. The genomic span covered by different technologies mentioned in this review.
Reads and optical maps derived from the NA12878 sample (DNA from a human individual sequenced as part of the 1000 Genomes Project) were mapped to the GRCh38 human genome reference. The histograms represent as follows: Illumina—the separation between natively generated paired-end reads (SRX1049855); Pacbio—the length of the reads generated by the Pacific Biosciences technology (SRX1607993); Oxford Nanopore—the length of the reads generated by the Oxford Nanopore technology (https://github.com/nanopore-wgs-consortium/NA12878); optical maps—the length of the fragments mapped by the BioNano nanocoding technology (from BioNano website); linked reads—the span of the region covered by reads originating from the same DNA fragment, as generated by the 10X Genomics technology (SRX1392293); Chicago—the separation between read pairs generated by the Chicago chromosome conformation capture protocol (SRX1423027); and Hi-C—the separation between read pairs generated by the Hi-C chromosome conformation capture protocol (SRX3651893).