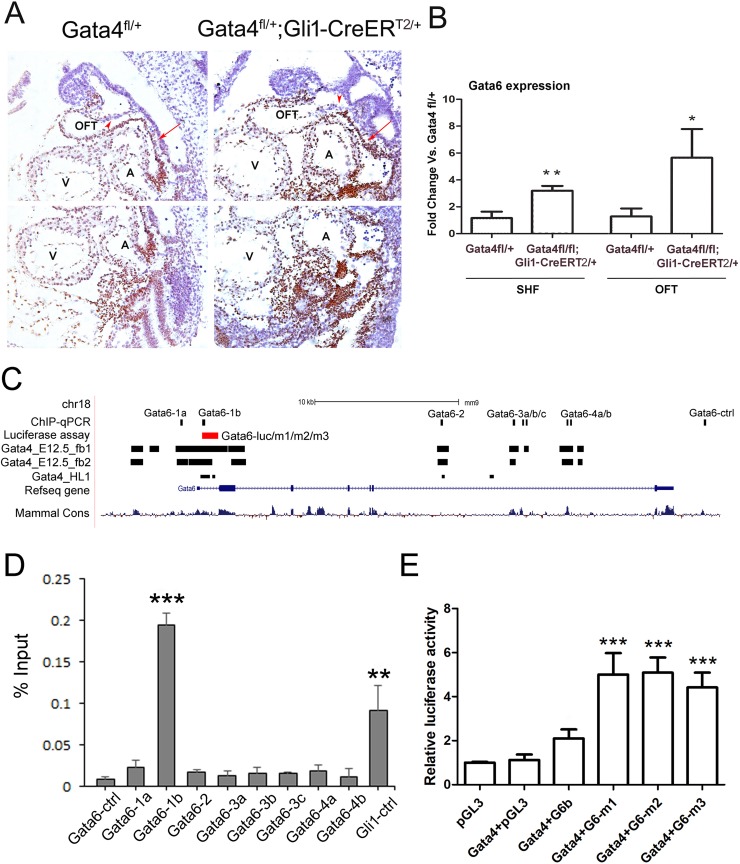
Fig 7. Gata6 is a repressing target of Gata4 in the SHF.
(A) IHC of the Gata6 in Gata4fl/+ and Gata4fl/+; Gli1-CreERT2/+ embryos at E9.5. The arrowhead indicated the NCCs-derived cells and the arrow indicates the splanchnic mesoderm. Magnification: 200X. (B) Gata6 was measured by realtime-PCR in the micro-dissected SHF and the OFT of the Gata4fl/+ and Gata4fl/fl; Gli1-CreERT2/+ embryos at E9.5. *p<0.1 vs. Gata4fl/+, **p<0.05, n = 3–5, compared with the expression in Gata4fl/+ embryos. OFT: out flow tract; A: atrium; V: ventricle. (C) Schematic of the mouse Gata6 genomic locus including Gata4-binding regions and the cloned genomic fragments used for Gata4 regulation assays (luciferase reporter assay and ChIP-qPCR). (D) Enrichment of Gata4-responsive Gata6 genomic fragments in the SHF by Gata4 ChIP-qPCR. Results are presented as mean ± SEM; n = 4; **P < 0.05 vs. Gata6-ctrl, ***P < 0.01, compared with Gata6-ctrl. (E) Gata4-stimulated firefly luciferase activity in wild-type Gata6 and Gata6 mutant fragments. Results are presented as mean ± SEM; n = 4; ***P < 0.01, compared with Gata4+pGL3.