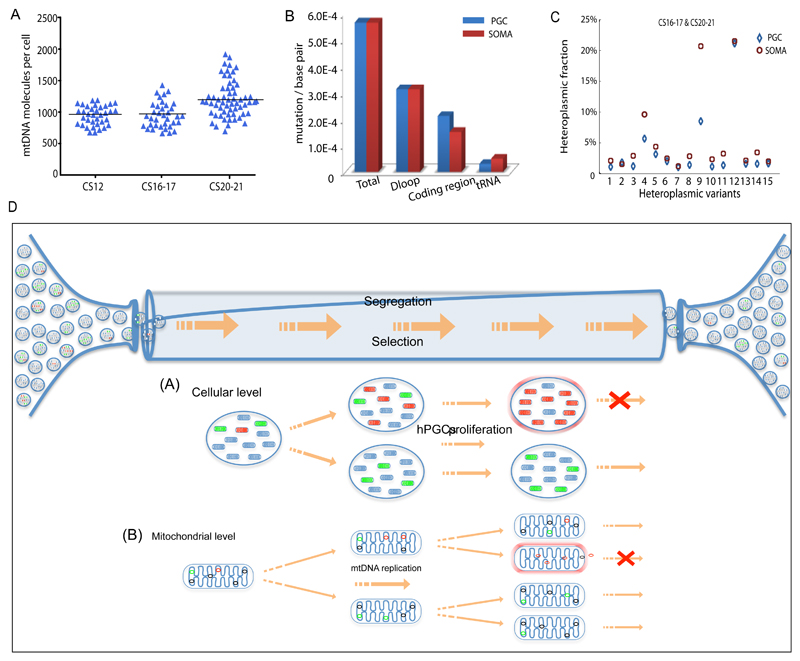
Figure 5. Mitochondrial DNA analysis of somatic cells in vivo & mechanisms of selection.
(a) MtDNA content of single human somatic cells in vivo at the same stage as the female primordial germ cells (PGCs) shown in Figs.1&2 (CS12 n=36, CS16-17 n=36; CS20-21 n=60). Each data point represents the mtDNA content in a single cell corresponding to the mean value from the independent qPCR measurements from 12 embryos. The horizontal bar represents the mean value.
(b) Mutation frequency for human female PGCs and corresponding somatic cells (soma) in developing human embryos (CS12-20/21) in vivo. Data derived from 12 embryos.
(c) Segregation of the same heteroplasmic mtDNA variants detected in somatic cells and PGCs from the same embryo at CS16-17 & CS20-21 (n = 15 variants) in vivo. Variants in the somatic cells had a higher heteroplasmy level than the PGCs (P=0.03, Fisher’s exact test, two-sided), which was not the case at CS12.
(d) In vivo PGCs contain ~ 300 discrete mitochondria which each containing 5 mtDNA molecules. In vitro data suggests that the amount of mtDNA may be even lower in female germ cells at an earlier stage. Some of the mtDNA molecules are mutated with either neutral (green) or deleterious (red) mutations. Between Carnegie Stage (CS) 12 and CS 20/21 the number of cells increases from ~3000 to ~80,000 accompanied by a massive proliferation of mtDNA molecules within the migrating PGCs. As the mitochondria replicate, the mutations will be copied. The random segregation of mtDNA molecules during mitochondrial fission will generate daughter mitochondria with either high or low levels of the original mutation. If the mutation is deleterious, then it will be selected against. This could be because it compromises ATP synthesis and activates surveillance systems that remove defective mitochondria, or because it compromises mtDNA replication, and thus prevents that particular mitochondrion from being copied. Both of these mechanisms can act in concert at the cellular level and at the mitochondrial level.