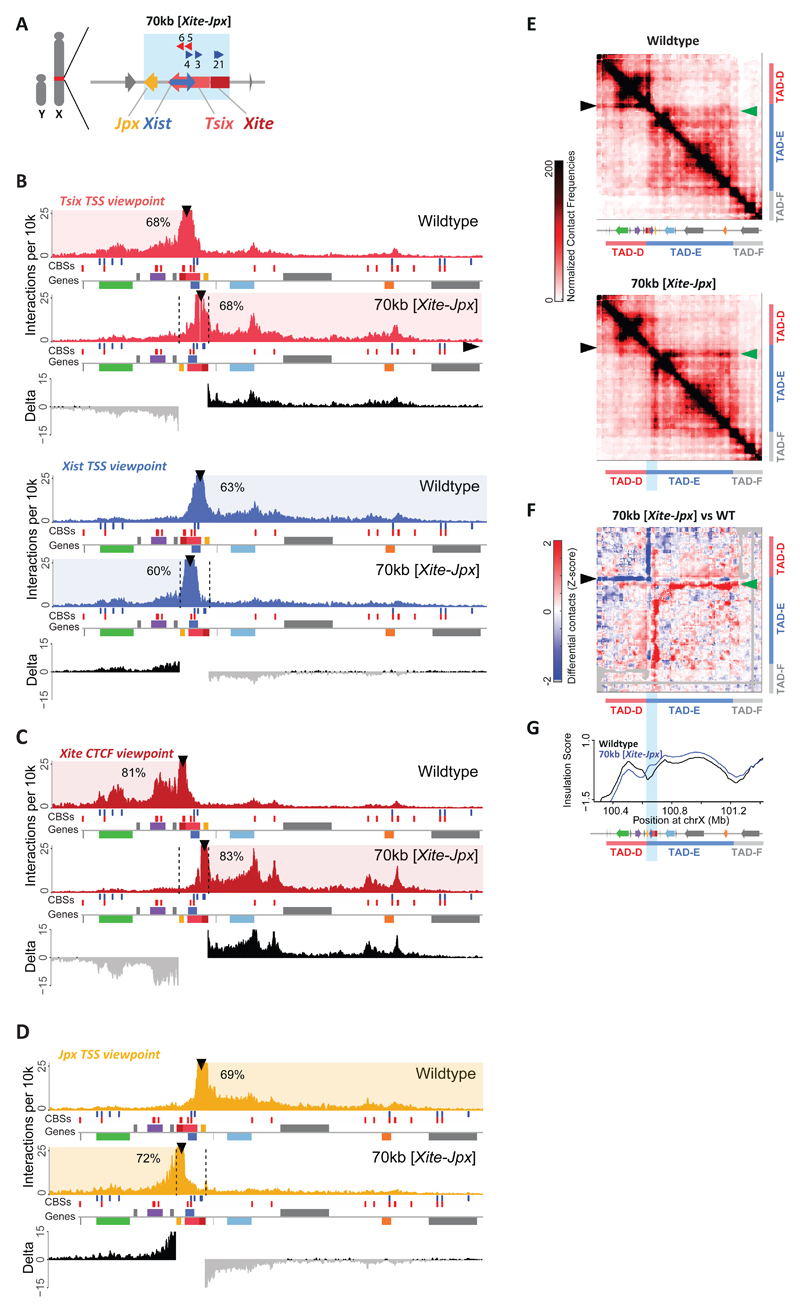
Fig. 2. Genomic inversion of the Xist/Tsix loci along with Xite and Jpx leads to topological changes within the Xic.
(a) Schematic representation of the 70-kb [Xite-Jpx] inversion in male mESCs. Blue box indicates inverted region. (b) Capture-C profiles and differential interaction frequencies, as in 1E, for Tsix (red) and Xist (blue) viewpoints in wild type and 70-kb [Xite-Jpx] inversion. (c) Capture-C profiles and differential interaction frequencies, as in 1E, for Xite (dark red) viewpoint in wild type and 70-kb [Xite-Jpx] inversion. (d) Capture-C profiles and differential interaction frequencies, as in 1E, for Jpx (yellow) viewpoint in wild type and 70-kb [Xite-Jpx] inversion. (e) 5C chromosome conformation contact frequencies in wildtype male mESCs (top) or harboring a 70-kb [Xite-Jpx] inversion (bottom). Pool of two replicates each, data for inversion have been inverted accordingly (i.e. map represents the inverted genome), normalized and binned as in 1C. Arrowheads indicate interactions described in the text. (f) Differential map represents the subtraction of wild type Z-scores from inversion Z-scores (see Methods). Gray pixels correspond to interactions that were filtered because they did not meet the quality control threshold (see Methods). Arrowheads indicate interactions described in the text. (g) Insulation scores, wild type in black and 70-kb [Xite-Jpx] inversion in blue. Inverted region represented by light blue box. Chromosome positions are aligned with panels E and F. Note: Each genomic alteration has been generated twice (two independent cell lines). Results for the first clone are shown here, results for the second clone are shown in Supplementary Figures 2-3.