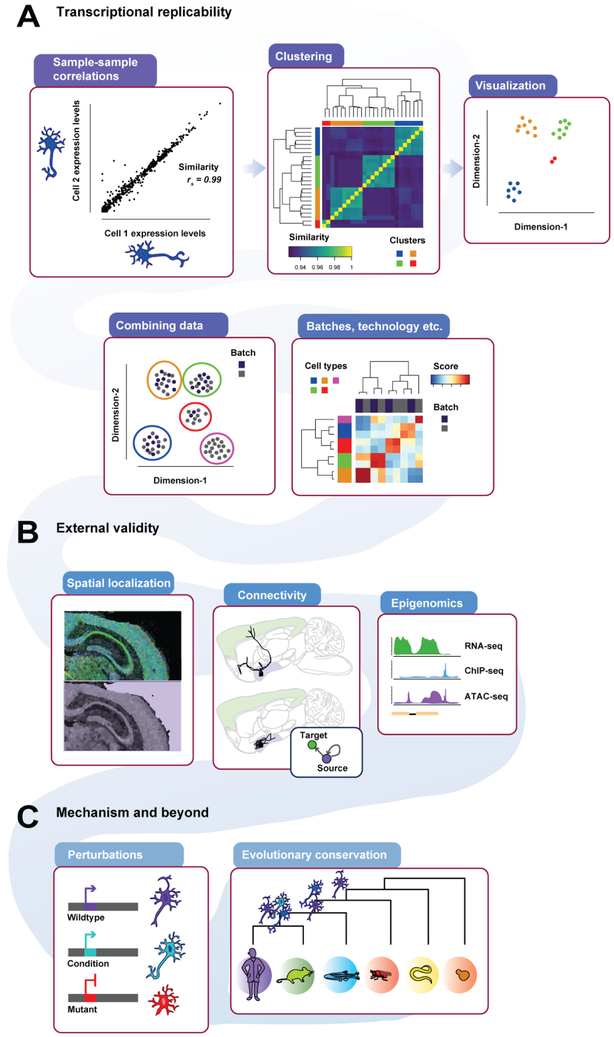
Figure 1 -. ScRNA-seq replicability from transcriptional profile to external validity to mechanism and beyond.
Expression profiles can be compared (A-top left) to characterize replicability or clustered (A-top middle) to find groups of similar expression profiles, also visible as groupings of cells (A-top right) when the data is summarized via dimension reduction. New datasets should show the same clusters (A-bottom left), which can then be assessed for similarity (A-bottom right). External validity may be partially established by examining spatial localization of markers (B-left), connectivity (B-middle) or epigenomic profiles (B-right). Studies of genetic perturbation (C-left) and conservation (C-right) provide insight into the molecular and evolutionary mechanisms that drive expression diversity.