Figure 2.
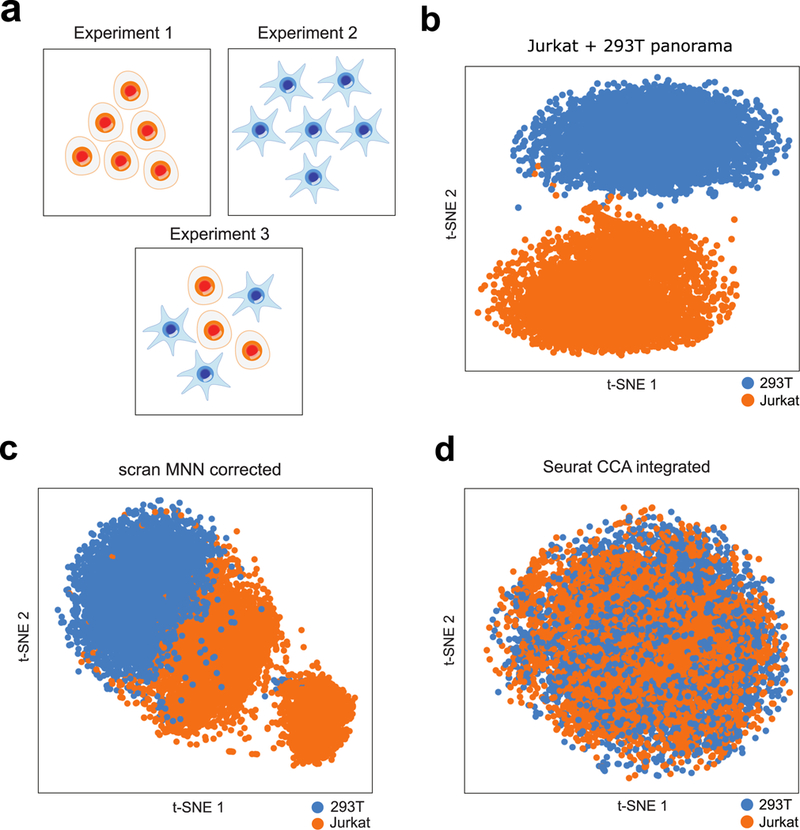
Scanorama correctly integrates a simple collection of datasets where other methods fail. (a) We apply Scanorama to a collection of three datasets17: one entirely of Jurkat cells (n = 3257 cells) (Experiment 1), one entirely of 293T cells (n = 2885 cells) (Experiment 2), and a 50:50 mixture of Jurkat and 293T cells (n = 3388 cells) (Experiment 3). (b) Our method correctly identifies Jurkat cells (orange) and 293T cells (blue) as two separate clusters. (c,d) Existing methods for scRNA-seq dataset integration are sensitive to the order in which they consider datasets (see Supplementary Fig. 1) and can incorrectly merge a Jurkat dataset and a 293T dataset together first before subsequently incorporating a 293T/Jurkat mixture, forming clusters that do not correspond to actual cell types.
