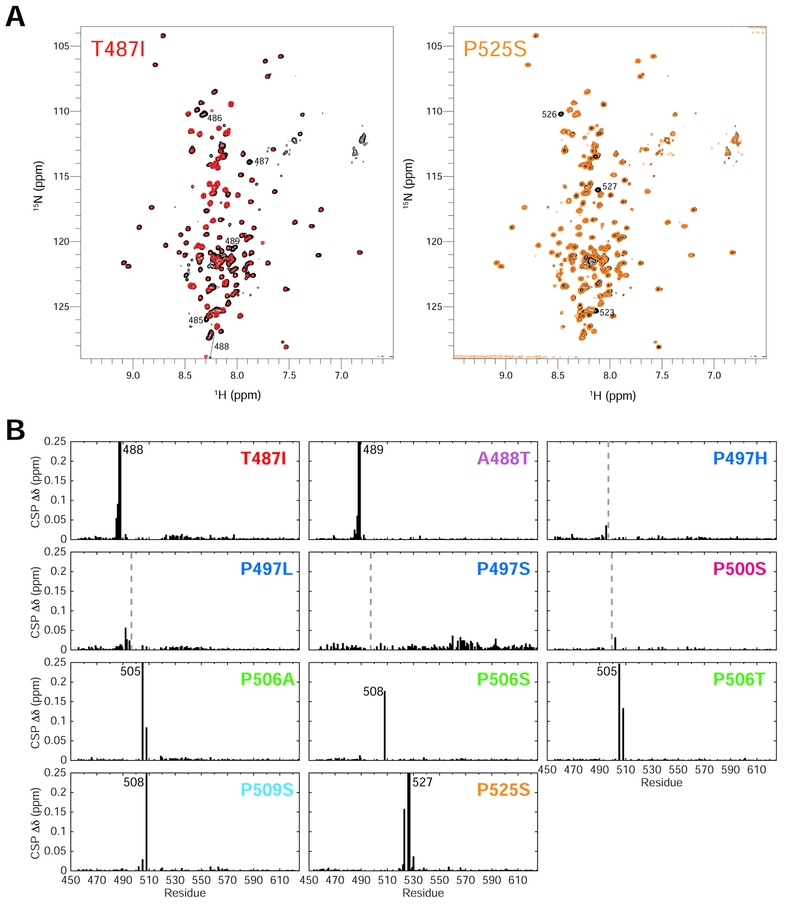
Figure 4. ALS-linked mutations minimally disrupt UBQLN2 structure.
(A) Representative NMR 1H-15N spectra of T487I (red) and P525S (orange) Pxx mutants compared to WT UBQLN2 (black). Spectra were collected under identical conditions, and contour settings are the same for all spectra. Those resonances with large chemical shift changes are labeled with residue number. (B) Backbone amide chemical shift perturbations (CSPs) are calculated between resonances of Pxx mutant and WT using 50 μM protein. CSPs are calculated as described in Methods. Dotted lines denote mutation site. In some cases, no CSPs were observed due to missing backbone assignments surrounding the mutation site.