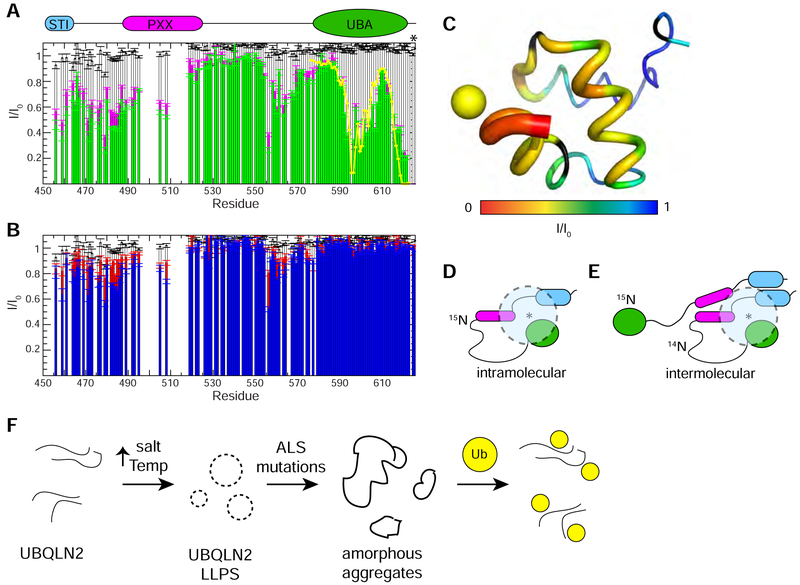
Figure 7. Ubiquitin disrupts interactions between UBA and Pxx region of UBQLN2.
(A) Intramolecular paramagnetic relaxation enhancement (PRE) effects on UBQLN2 450–624 (50 μM) in the absence of Ub (green) and presence of Ub (magenta) with MTSL spin label attached to an engineered Cys at residue 624 (marked with an asterisk). Yellow line represents back-calculated PREs derived from determining the position of MTSL’s unpaired electron in the UBA structure (see part C). White bars represent PRE effects in a control 50 μM sample with unconjugated MTSL in parts A and B. (B) Intermolecular PREs on WT UBQLN2 450–624 (25 μM) in a 1:1 mixture with NMR-invisible UBQLN2 450–624 S624C (25 μM) with MTSL spin label attached. Intermolecular PREs in the absence of Ub (blue) and presence of Ub (red). Errors in I/I0 for (A) and (B) were determined using the standard error propagation formula for ratios. (C) PRE effects from MTSL are mapped onto the structure of UBQLN2 UBA via backbone thickness and color coding using PDB: 2JY6. Yellow sphere represents back-calculated position of MTSL’s unpaired electron. (D,E) Representative schematic model of results from intramolecular (D) and intermolecular (E) PRE experiments whereby spin label is represented by an asterisk, and dotted semi-transparent circle represents residues’ amide resonances that are attenuated by spin label. (F) Proposed model of how ALS-linked mutations disrupt UBQLN2 LLPS.