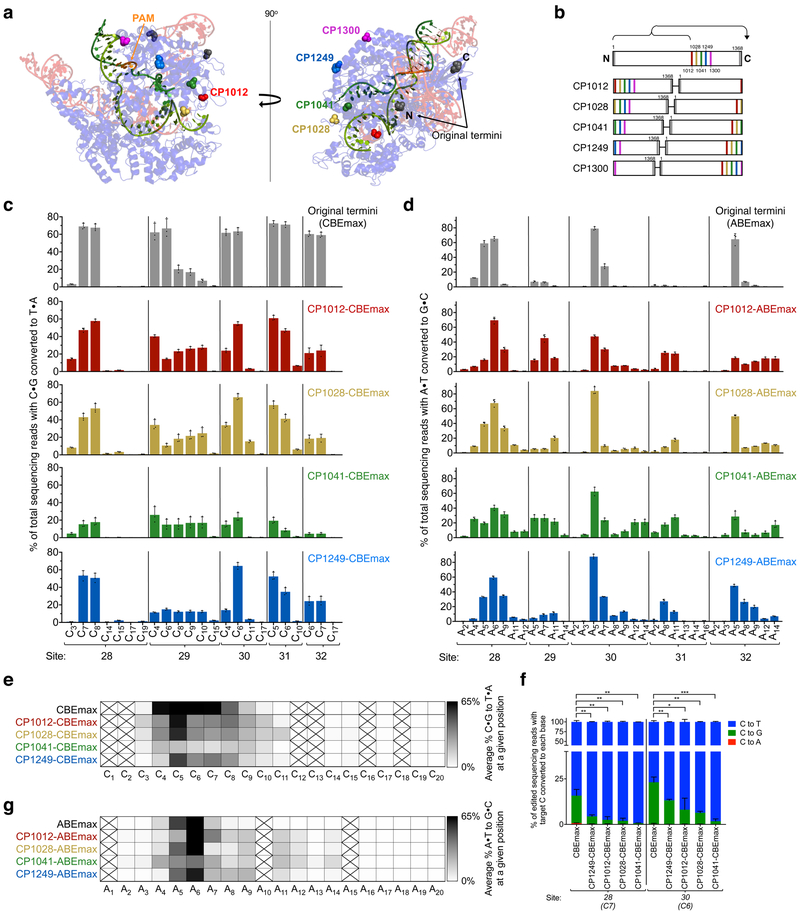
Figure 2. Circularly permuted Cas9 variants are compatible with the cytosine and adenine base editor architectures and exhibit broadened editing windows.
(a) Two views of the SpCas9:sgRNA:DNA crystal structure (PDB: 5F9R36) showing the location of the N and C termini in the wild-type protein (black) and in circularly permuted variants tested in this work (red, yellow, green, blue, and magenta). The DNA strand paired with the guide RNA is shown in light green and the other strand, targeted for base editing and partially disordered in the structure, is shown in dark green. (b) Schematic representations of CP-Cas9 variants tested in this work, numbered to indicate the original SpCas9 amino acid that serves as the new N-terminus. (c) Base editing in HEK293T cells by CP-CBEmax variants across five genomic sites. (d) Base editing in HEK293T cells by CP-ABEmax variants across five genomic sites. For clarity, in (c) and (d), protospacer positions with editing lower than 0.5% across all editors are not shown. CP1300 data is shown in Supp. Fig. 4. (e, f) Heat maps showing average base editing efficiency at each position within the protospacer across five sites tested with (e) CP-CBEmax variants or (f) CP-ABEmax variants, normalized to the maximum observed editing within the protospacer (1.0). Boxes crossed out indicate positions for which no target base was present among all genomic sites tested. (g) The product distribution among edited DNA sequencing reads (reads in which the target C is base edited) is shown for each CP-CBEmax variant tested at two different genomic sites that are especially prone to non-C-to-T byproduct formation. Subscripted numbers indicate protospacer positions, counting the first base of the PAM as position 21. Values and error bars reflect the mean±s.d. of three biological replicates performed on different days at each site. ns, P>0.05; *P<0.05; **P<0.01; ***P<0.001, by two tailed Student’s t-test.