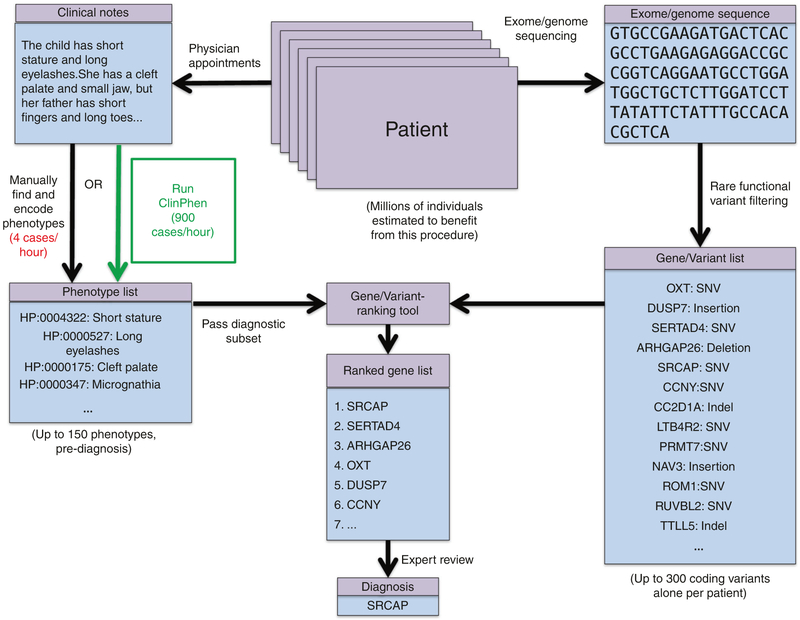
Fig. 1. Steps to diagnose a patient with a Mendelian disease using automated gene-ranking algorithms.
The patient’s genotypic information is encoded using standard formats (variant call format [VCF] file, candidate causative gene list) and a list of patient phenotypes encoded as ontology terms. Extensive tool support exists for obtaining candidate causative variants and genes from an exome sequence. Tool support for obtaining an appropriate list of encoded patient phenotypes from the patient’s clinical notes is limited. Encoded phenotypes are currently acquired by manually reading through the patient’s clinical notes and recording the phenotypes found as their IDs in a phenotype ontology. We introduce ClinPhen, a tool that automates phenotype extraction from clinical notes, optimized to accelerate patient diagnosis. SNV single-nucleotide variant.