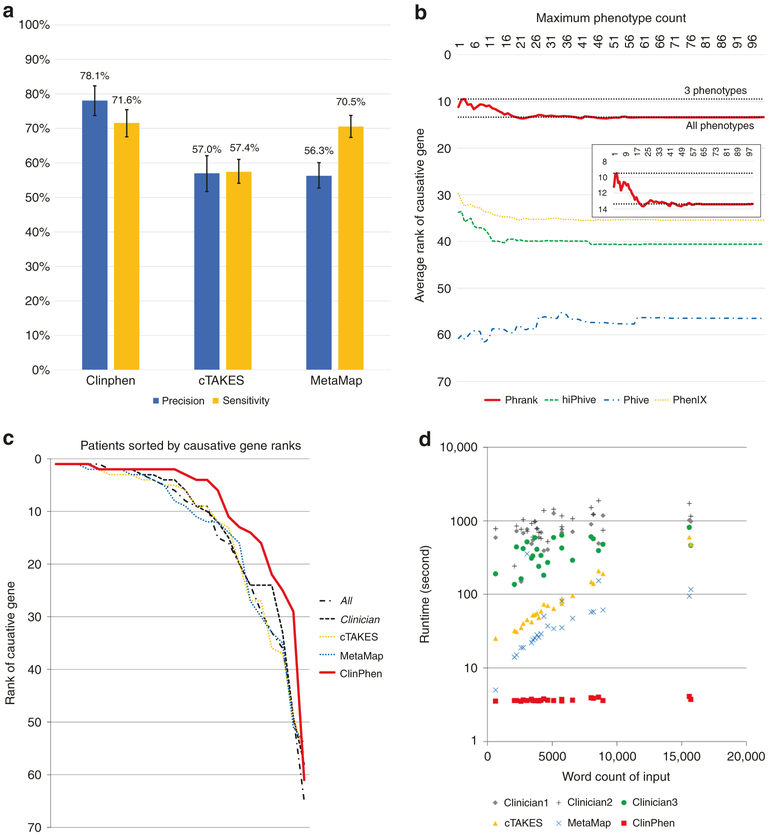
Fig. 3. Performance of all extraction methods.
(a) Comparison of the extractors’ precision and phenotype sensitivity (higher bars mean higher accuracy). We compared the average precision and sensitivity values of ClinPhen, cTAKES, and MetaMap, using patients from the Stanford test set as subjects, and the All set (all of the phenotypes found manually and confirmed by a physician to apply to the patient) as the correct phenotypes. The average (column) and 95% confidence interval (calculated using bootstrapping with 1000 trials) of the precision and sensitivity values across all patients are displayed for each extractor. ClinPhen achieves the highest average precision and sensitivity. (b) Causative gene-ranking performance of each gene-ranking tool when run with different numbers of phenotypes returned by ClinPhen (lower number means better causative gene rankings). ClinPhen was run on the clinical notes of the Stanford test set, and the gene-ranking tools were called with the patient’s genetic information and the n highest-priority (most-mentioned, first-occurring) extracted phenotypes, with n running from 1 to 100 inclusive. The average causative gene rank across all patients was taken for each phenotype count limit (n)/gene-ranking tool pairing. The better-performing gene-ranking algorithms rank the causative gene higher when run with a few (around 3) high-priority phenotypes than with all extracted phenotypes. (c) Phrank’s causative gene-ranking performance across all extraction methods (lower numbers mean better causative gene rankings). We compared the causative gene ranks obtained by running Phrank on the Stanford test set with various extracted sets of phenotypes (All manually found, physician-verified phenotypes [All] versus a subset of phenotypes considered by a physician to be useful for diagnosis [Clinician] versus automatically extracted phenotypes using various methods). Phrank ranks are sorted lowest to highest for each extractor. Phrank performs better when run with ClinPhen’s 3 highest-priority phenotypes (the most-mentioned, earliest-occurring phenotypes in a patient’s clinical notes) than when run with other phenotype sets, manually or automatically extracted. (d) Extractor runtime comparison on each patient (lower number means faster runtime). We measured the runtime of each extractor (ClinPhen, cTAKES, and MetaMap) on each patient’s clinical notes, in seconds. For each patient, we also measured the time three clinicians took to manually scan through the same notes read by the automatic extractors, and encode the phenotypes considered useful for diagnosis. Each data point is one patient whose clinical notes were scanned by one of the extractors (or clinicians). The horizontal position is the total number of words in the patient’s clinical notes. The vertical position is the time taken for the extractor to run on the notes (logarithmically scaled). While MetaMap’s runtime scales linearly and cTAKES’ runtime scales exponentially with the total length of the clinical notes, ClinPhen runs in near-constant time, and is 15–20× faster than the next fastest tool. All automatic extraction tools are much faster than manual extraction.