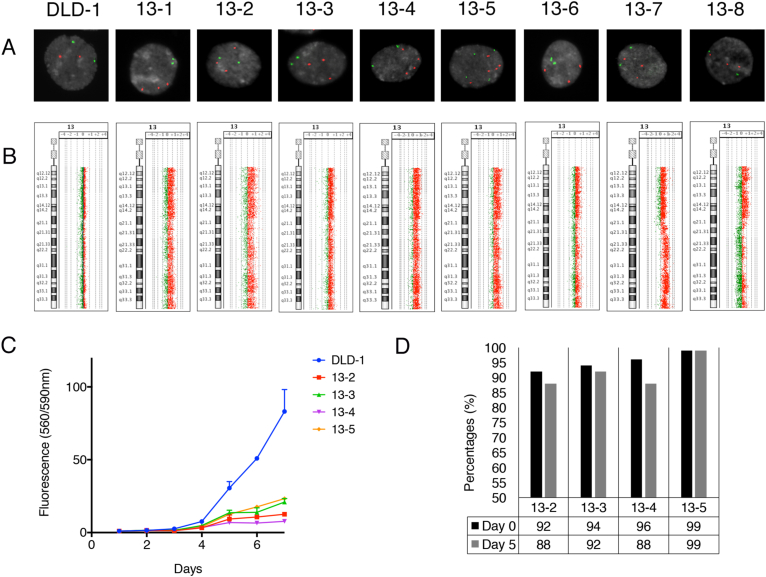
Figure 1.
Characterization of the eight single cell derived clones of DLD-1 + 13 cells. (A) FISH probes corresponding to the MET gene (chromosome band 7q31, green signals) and CDX2 (chromosome band 13q21, red signals) hybridized onto interphase nuclei for the parental DLD-1 WT and the eight DLD-1 + 13 clones. One hundred fifty cells were counted per clone. (B) Genomic aberration profile in the DLD-1 WT clone and all eight DLD-1 + 13 clones using aCGH. (C) Proliferation graph for the DLD-1 WT and four DLD-1 + 13 clones. (D) Percentages of cells (n = 100) with a gain of chromosome 13 as analyzed by FISH for four DLD-1 + 13 clones on day 0 and day 5 without neomycin selection. The FISH probes used were MET (7q31) and CDX2 (13q12). A gain of chromosome 13 was made if there were more copies of CDX2 as compared to MET within the same cell.