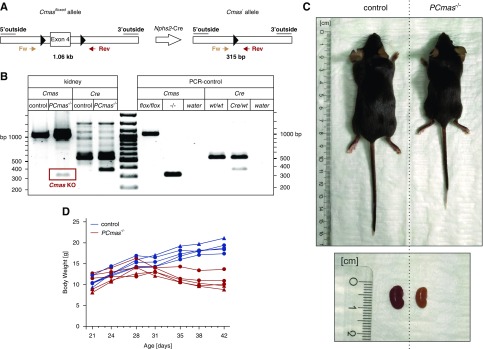
Figure 1.
Podocyte-specific CMAS-knockout (PCmas−/−) mice show altered physiognomy and body weight loss. (A) Targeting strategy. Exon 4 of Cmas was flanked by loxP sites (Cmasfloxed). Podocyte-specific deletion of Cmas was achieved by expressing Cre recombinase under control of the Podocin (Nphs2) promoter. Forward (Fw) and reverse (Rev) primers for Cmas PCR analysis and product sizes are indicated. (B) Genotypic characterization of control and PCmas−/− mice. Genotypes were analyzed by PCR using genomic DNA isolated from kidney biopsy specimens sampled at P43 by the Fw/Rev primer pair (see [A]), enabling amplification of the floxed (1.06 kb) as well as the deleted (315 bp) Cmas allele (left-hand side of the ruler; Cmas− allele is highlighted in a red box). The PCR to amplify the Cre allele (372 bp) was supplemented with a control primer pair (PST Fw/PST Rev) to confirm DNA quality, giving rise to a 530 bp fragment. Specificity of PCR reactions (“PCR control”) was confirmed by using genomic DNA from mouse tail biopsy samples as template. Murine Cmas−/− ES cells (−/−) were used as template to demonstrate the size of the Cmas− allele (“PCR control”). (C) Physiognomy of PCmas−/− and littermate control mice (top) and kidneys (bottom) at P46. (D) Time course of body weight of control (blue, n=5) and PCmas−/− (red, n=5) mice. Circle, female; triangle, male.