Abstract
Background
Elevated protein expressions of CD markers such as IL2RA/CD25, CXCR4/CD184, CD34 and CD56 are associated with adverse prognosis in acute myeloid leukemia (AML). However, the prognostic value of mRNA expressions of these CD markers in AML remains unclear. Through our pilot evaluation, IL2RA mRNA expression appeared to be the best candidate as a prognostic biomarker. Therefore, the aim of this study is to characterize the prognostic value of IL2RA mRNA expression and evaluate its potential to refine prognostification in AML.
Methods
In a cohort of 239 newly diagnosed AML patients, IL2RA mRNA expression were measured by TaqMan realtime quantitative PCR. Morphological, cytogenetics and mutational analyses were also performed. In an intermediate-risk AML cohort with 66 patients, the mRNA expression of prognostic biomarkers (BAALC, CDKN1B, ERG, MECOM/EVI1, FLT3, ID1, IL2RA, MN1 and WT1) were quantified by NanoString technology. A TCGA cohort was analyzed to validate the prognostic value of IL2RA. For statistical analysis, Mann–Whitney U test, Fisher exact test, logistic regression, Kaplan–Meier and Cox regression analyses were used.
Results
In AML cohort of 239 patients, high IL2RA mRNA expression independently predicted shorter relapse free survival (RFS, p < 0.001) and overall survival (OS, p < 0.001) irrespective of age, cytogenetics, FLT3-ITD or c-KIT D816V mutational status. In core binding factor (CBF) AML, high IL2RA mRNA expression correlated with FLT3-ITD status (p = 0.023). Multivariable analyses revealed that high IL2RA expression (p = 0.002), along with c-KIT D816V status (p = 0.013) significantly predicted shorter RFS, whereas only high IL2RA mRNA expression (p = 0.014) significantly predicted shorter OS in CBF AML. In intermediate-risk AML in which multiple gene expression markers were tested by NanoString, IL2RA significantly correlated with ID1 (p = 0.006), FLT3 (p = 0.007), CDKN1B (p = 0.033) and ERG (p = 0.030) expressions. IL2RA (p < 0.001) and FLT3 (p = 0.008) expressions remained significant in predicting shorter RFS, whereas ERG (p = 0.008) and IL2RA (p = 0.044) remained significant in predicting shorter OS. Similar analyses in TCGA intermediate-risk AML showed the independent prognostic role of IL2RA in predicting event free survival (p < 0.001) and OS (p < 0.001).
Conclusions
High IL2RA mRNA expression is an independent and adverse prognostic factor in AML and specifically stratifies patients to worse prognosis in both CBF and intermediate-risk AML.
Electronic supplementary material
The online version of this article (10.1186/s12967-019-1926-z) contains supplementary material, which is available to authorized users.
Keywords: Acute myeloid leukemia, Prognosis, IL2RA, mRNA expression, Core binding factor AML, Intermediate-risk AML, NanoString
Background
Acute myeloid leukemia (AML) is a well-known heterogeneous hematological malignancy with a broad range of prognosis, which is greatly impacted by clinical factors, cytogenetics and molecular characteristics [1]. In the current risk stratification system, recurrent genetic abnormalities stratify AML into three risk status categories including favourable-risk, intermediate-risk and poor-risk [1]. This risk stratification system, together with clinical characteristics of AML patients such as age and medical comorbid, dictates the prognosis of each individual patient, as well as guide physicians to decide appropriate treatment regimens [1]. For instance, in younger adult patients high-dose cytarabine-based therapy has been recommended as the conventional consolidation regimen for AML of favourable-risk, whereas allogeneic hematopoietic stem cell transplantation (HCT) for AML of poor-risk [1]. Nevertheless, either regimen can be considered for AML of intermediate-risk [1]. While patients have benefited greatly from current risk stratification strategies of AML, the prognosis of AML in each risk category is still quite variable. Further improvement of prognostic tools is needed to better stratify these patients and guide treatments accordingly. In addition to the well-established recurrent cytogenetic aberrations and molecular mutations, genes with aberrant expression at protein or mRNA level have also been shown to have significant prognostic values in AML over the last decade [2]. These gene expression biomarkers not only help shed light on mechanisms of development and progression of this largely heterogeneous malignancy, but more importantly help clinicians to refine prognostic tools to improve patient care in clinical practice.
Elevated protein expressions of various cluster of differentiation (CD) marker genes such as interleukin 2 receptor subunit alpha (IL2RA/CD25) [3–7], C-X-C chemokine receptor type 4 (CXCR4/CD184) [8–10], CD34 [11, 12] and CD56 [13, 14] have been shown to predict poor clinical outcome in AML. Most studies used flow cytometry (FCM) technology to quantify protein expression of these CD markers. Meanwhile, many studies based on mRNA quantification platforms identified mRNA expression biomarkers in AML, such as BAALC, ERG, MECOM/EVI1 and WT1, which could offer additional prognostic values to improve current stratification system [15–21]. RNA quantitative methodology, in comparison to FCM method, have several unique advantages. Firstly, stored RNA or bone marrow (BM) samples could be used to accurately quantify RNA gene expression when fresh samples are not available. This enhanced tissue flexibility would potentially improve patient care in practice. In addition, this approach makes retrospective analysis of samples feasible, which also enhance clinical care and facilitate research. Secondly, only limited tissue is needed for RNA quantitation that could significantly improves tissue efficiency. Lastly, many RNA quantitative platforms are designed to perform multiplex gene testing to improve consistency and efficiency.
By far, the prognostic values of mRNA expression of most CD markers in AML remain elusive. To our knowledge, the only prognostic CD marker, of which the mRNA expression prognostic value has been indicated, is CD34 [22, 23]. However, reports on prognostic values of CD34 mRNA and protein expressions were not consistent [24–26]. In general, gene expression levels at mRNA level do not necessarily correlate well with those at protein level [27, 28] which are subject to multiple layers of regulation [27–29]. In addition, the protein levels of known CD biomarkers in AML are generally quantitated by FCM assay on blast cells, whereas mRNA expression levels of CD markers are quantitated in bulk tissue using different platforms. Therefore, it is important to investigate and validate the prognostic value of these CD biomarkers at mRNA level independently.
In our study, we initially sought to investigate the prognostic value of mRNA expressions of various CD biomarkers in AML and study if they can add independent prognostic value to the established prognostic factors. We conducted a pilot study to evaluate correlations of mRNA/protein expressions of four prognostic CD marker genes (IL2RA, CXCR4, CD34 and CD56), analyzed the prognostic values of their mRNA expressions in the TCGA-LAML cohort and determined IL2RA as best candidate gene for further study in larger cohort. Subsequently, in our clinical cohorts, we aimed to systemically evaluate the prognostic value of IL2RA mRNA expression in AML in the context of clinical and laboratory factors with prognostic relevance. We further characterized its prognostic role in core binding factor (CBF) AML in the context of established prognostic factors, as well as in intermediate-risk AML in the context of other mRNA expression prognostic factors (ERG, ID1, WT1, FLT3, WT1, BAALC, CDKN1B, MECOM/EVI1, MN1). Our study not only consistently reveals independent prognostic value of mRNA expression of IL2RA in AML, particularly in CBF and intermediate-risk AML, but also serves as a proof-of-concept study for future research endeavors to investigate prognostic roles of mRNA expression of other CD biomarkers.
Methods
Patients and treatments
We analyzed mRNA expression of IL2RA using BM samples from 239 adult patients (age range: 15–65) diagnosed with AML between 2012 and 2016 at Institute of Hematology, Wuhan Union Hospital. The patients received intensive induction chemotherapy and consolidation chemotherapy or HCT [1]. Cases of acute promyelocytic leukemia (APL) were not included in this cohort. We also analyzed expression of IL2RA together with eight other known prognostic genes simultaneously in a multigene panel testing platform by NanoString using BM samples from 66 adult patients diagnosed with intermediate-risk AML. The diagnosis of AML was made according to World Health Organization classification [30] and French–American–British (Fab) classification. Cytogenetic risk stratification was defined according to the AML NCCN guideline version 3. 2017. This study was approved by the ethics committee of Tongji Medical College, Huazhong University of Science and Technology and was carried out in accordance with the Helsinki Declaration.
Cytogenetic analysis
Conventional cytogenetic analysis was performed on G-banded preparations from 48-h bone marrow cell cultures. The chromosomal aberrations were described according to the International System for Cytogenetic Nomenclature (ISCN) 2009 [31].
Flow cytometry
Flow cytometry analysis on fresh bone marrow samples were carried out at the time of diagnosis. In addition to monoclonal antibodies (Abs) and isotype control IgGs used for diagnostic immunophenotyping [32], antibodies for IL2RA/CD25 (347643, BD Bioscience, CA) and CXCR4/CD184 (555976, BD Bioscience Pharmingen, CA) were used. Flow cytometry analyses were performed as previously described [32].
DNA extraction and mutational analyses
Genomic DNA was extracted from diagnostic marrow specimens BM samples using standard method and FMS-like tyrosine kinase 3-internal tandem duplication (FLT3-ITD) [33], nucleophosmin 1 (NPM1) [34], CEBPA [35] and c-KIT D816V [36] mutation status were evaluated as previously described. Direct sequencing of DNA was performed using Thermo Sequence Dye Terminator sequencing reaction and ABI Prism 3730 sequencing analyzer (Thermo Fisher Scientific, Carlsbad, CA).
RNA extraction and TaqMan RQ-PCR
BM samples RNA extraction were performed as previously described [37]. Reverse transcription and cDNA synthesis were prepared from ~ 1.0 μg of total RNA using the PrimeScript RT Master Mix (Takara Bio Inc, Japan) according to the manufacturer’s instruction and 1/8 of the cDNA was used as a template for each PCR reaction. PCR reactions and fluorescence quantitative measurements were conducted on the Applied Biosystems Prism 7500 instrument (PE Applied Biosystems, USA). TaqMan Fast Advanced Master Mix (ThermoFisher Scientific, USA) were used to perform the TaqMan assay according to the manufacturer’s instruction. The IL2RA gene expression was measured by TaqMan assay Hs00907779_m1 and the CXCR4 gene expression was measured by TaqMan assay as previously described [38]. ABL1 gene was selected as the reference gene to compensate for the variations in mRNA and cDNA, and the ABL1 TaqMan assay was described in previous study [39]. The comparative cycle threshold (Ct) method was used to calculate the relative expression of target genes [40]. The threshold cycles for IL2RA, CXCR4 and ABL1 were determined by replicates and the mean CT was used for calculation. The cycle number difference was calculated (ΔCt = Ct target gene − CtABL1) and the relative expression values of target gene were expressed as 2−ΔCT. The PCR efficiencies of IL2RA and ABL1 assays have been evaluated by standard curves. The standard curves for ABL1 assays were generated by tenfold dilution series of four different plasmid concentrations (copy numbers 103; 104; 105; 106) and the standard curves for IL2RA assays were generated by tenfold dilution series of positive RNA samples (1; 1:10; 1:102; 1:103). The PCR efficiencies calculated for either ABL1 or IL2RA assays varied between 95.0 and 105.0%. Negative controls and interassay positive controls have been included in the PCR assays. The data were analyzed using the Applied Biosystems 7500 software v2.0.5.
NanoString nCounter assay
The probes sequences for the prognostic transcripts including the reference gene ABL1 were designed according to principles described before [41]. Six positive and eight negative spike-in control probes were included in the probes codeset and were used for normalization of raw data. The counts of the reference gene ABL1 were used to further normalize the gene counts. For each NanoString assay, about 250 ng sample RNA was used for each reaction. The experimental procedures were carried out on the NanoString Preparation Station and Digital Analyzer according to manufacturer’s instructions [37, 41]. The nSolver 3.0 software were used for analyses.
TCGA dataset
Level 3 RNA-seq gene expression data (fragments per kilobase per million mapped fragments/FPKM values) of 151 AML cases were retrieved from the TCGA data portal and clinical data from the cohort were retrieved from Additional file 1: Table S1 of the publication of the TCGA-LAML study [42]. Of the 151 cases, there are 136 cases of AML cases excluding APL and 80 cases of intermediate-risk AML.
Statistical analysis
Mann–Whitney U test or Kruskal–Wallis test was used for comparison of continuous variable between different subsets of patients. Fisher exact or Chi-square tests were used for comparison of categorical variables between different subsets of patients. The bivariate Spearman correlation test was used to measure associations among continuous variables. Normalized NanoString counts were log2 transformed before the correlation analysis. Complete remission (CR), RFS and OS were defined as previous described [43]. Univariable logistic regression models were performed to evaluate the achievement of CR. Univariable survival analyses using Kaplan–Meier method were performed to evaluate association of RFS or OS for dichotomous variables, the differences were calculated by log-rank test. Univariable survival analyses for continuous variables was performed using Cox regression method. Multivariable logistic regression models were generated for CR and multivariable Cox regression models for RFS and OS. The statistical analyses were performed using IBM SPSS 19.0.0.
Results
The pilot study revealed the mRNA expressions of IL2RA is the most suitable prognostic CD marker gene
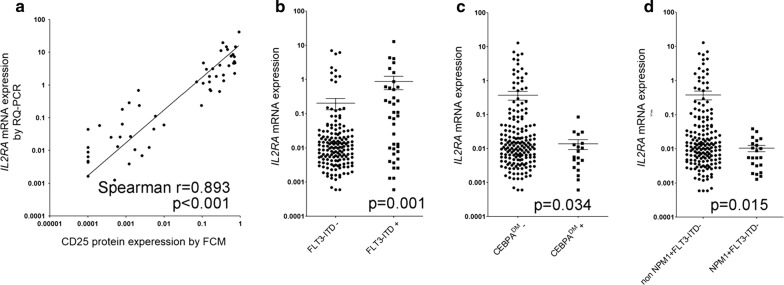
In order to determine the suitable prognostic biomarker candidate genes, we performed initial evaluation of both mRNA as well as protein levels on four CD marker genes (IL2RA, CXCR4, CD56 and CD34). The IL2RA mRNA and its protein CD25 levels were assayed in 54 primary AML BM samples by TaqMan RQ-PCR and FCM respectively, in which the mRNA level was shown as 2−∆Ct of IL2RA gene relative to ABL1 gene and the expression level of CD25 protein was illustrated as percentage in leukemia blast cells. A significant association was found between mRNA expression of IL2RA gene and protein expression of CD25 (p < 0.001 and correlation coefficient Spearman r = 0.893, Fig. 1a and Additional file 1: Table S1). Although the percentage of leukemia blasts varies from 20 to 99% in the diagnostic bone marrow specimens of this study group, IL2RA expression level was independent of the blast percentage (p = 0.626, Spearman r = − 0.0679, Additional file 1: Fig. S1). The CXCR4 mRNA and its protein CD184 expressions were examined in 41 adult AML patients by TaqMan RQ-PCR and FCM, whereas no significant association was found (p = 0.863 and Spearman r = 0.0278, Additional file 1: Fig. S2 and Table S1). Correlations between mRNA and protein expressions of CD34 and CD56 genes have been studied and reported previously in two independent studies using leukemia samples [44, 45] (Additional file 1: Table S1). Significant correlation of CD34 mRNA/protein expressions has been shown in both studies, whereas inconsistent results were indicated for CD56 (p < 0.01 and p = 0.180 respectively, Additional file 1: Table S1).
Fig. 1.
Correlation of mRNA/protein expressions of IL2RA and IL2RA mRNA expression levels according to mutational status. a Correlation of IL2RA mRNA expression with its protein expression is significant, p < 0.001 and correlation coefficient Spearman r = 0.893. b There is much higher level of IL2RA and in FLT3-ITD+ group than in FLT3-ITD− group, p = 0.001. c There is much lower level of IL2RA mRNA expression and in CEBPADM group than in non-CEBPADM group, p = 0.034. d There is much lower level of IL2RA mRNA expression in NPM1+FLT3-ITD− group than in nonNPM1+FLT3-ITD− group, p = 0.015
Next, we performed univariable Cox regression analysis to evaluate prognostic role of mRNA expressions of the four CD markers genes as continuous variables on OS in an existing TCGA-LAML cohort (2013). Among these CD markers, only the mRNA expression of IL2RA/CD25 seemed to significantly predict OS in AML (p < 0.001, Additional file 1: Table S1). Therefore, we selected mRNA expression of IL2RA as the best suitable candidate prognostic biomarker for further validation analysis in a larger AML cohort.
Patients characteristics of AML cohort and association with IL2RA mRNA expression
In a cohort of 239 newly diagnosed AML adult patients, we quantified the IL2RA mRNA expression using BM RNA samples by TaqMan RQ-PCR. The characteristics of patients including clinical, cytogenetics and molecular features in this cohort were described and their correlation with IL2RA mRNA expression level were analyzed and summarized in Table 1. Significant differences of IL2RA mRNA expression were observed in AML with certain molecular abnormalities (Table 1). In particular, a positive correlation of IL2RA mRNA expression level with FLT3-internal tandem duplication (FLT3-ITD) mutation (p = 0.001, Table 1 and Fig. 1b) and a negative correlation with CEBPA double mutations (CEBPADM) (p = 0.034, Table 1 and Fig. 1c) or NPM1 mutation in the absence of FLT3-ITD(NPM1+FLT3-ITD−, p = 0.015, Table 1 and Fig. 1d) were found.
Table 1.
Correlation of IL2RA mRNA expression with clinical and laboratory parameters in AML cohort
| N | Percentage (%) | IL2RA mRNA expression range | Median |
IL2RA mRNA expression Mean ± SE |
p-value | |
|---|---|---|---|---|---|---|
| Total | 239 | |||||
| Age group | 0.312 | |||||
| 15–29 | 65 | 27.2 | 0.0014–3.780 | 0.0128 | 0.211 ± 0.603 | |
| 30–49 | 113 | 47.3 | 0.0006–7.056 | 0.0097 | 0.304 ± 1.092 | |
| 50–65 | 61 | 25.5 | 0.0013–12.870 | 0.0130 | 0.598 ± 2.184 | |
| Gender | 0.973 | |||||
| Male | 127 | 53.1 | 0.0006–12.870 | 0.0105 | 0.395 ± 1.573 | |
| Female | 112 | 46.9 | 0.0006–6.140 | 0.0122 | 0.306 ± 1.108 | |
| Blast percentage (%) | 0.941 | |||||
| ≥ 50 | 149 | 62.3 | 0.0006–8.540 | 0.0113 | 0.349 ± 1.247 | |
| 20–49 | 90 | 37.7 | 0.0006–12.870 | 0.0114 | 0.361 ± 1.566 | |
| WBC count | 0.684 | |||||
| ≥ 30 billion/L; high | 58 | 34.7 | 0.0007–5.890 | 0.0136 | 0.341 ± 1.060 | |
| < 30 billion/L; low | 109 | 65.3 | 0.0006–12.870 | 0.0106 | 0.430 ± 1.649 | |
| AML type | 0.605 | |||||
| De novo AML | 231 | 96.7 | 0.0006–12.870 | 0.0113 | 0.353 ± 1.392 | |
| Secondary AML | 8 | 3.3 | 0.0033–1.560 | 0.0258 | 0.365 ± 0.642 | |
| Fab subtypes | 0.279 | |||||
| Fab M0 | 6 | 2.6 | 0.0026–0.0468 | 0.0123 | 0.0172 ± 0.0161 | |
| Fab M1 | 32 | 13.6 | 0.0013–6.140 | 0.0136 | 0.402 ± 1.474 | |
| Fab M2 | 118 | 50.2 | 0.0006–4.120 | 0.0096 | 0.146 ± 0.576 | |
| Fab M4 | 43 | 18.3 | 0.0007–12.870 | 0.0175 | 0.605 ± 2.097 | |
| Fab M5 | 26 | 11.1 | 0.0006–8.540 | 0.0060 | 0.689 ± 1.981 | |
| Fab M6 | 10 | 4.3 | 0.0020–7.056 | 0.0521 | 0.749 ± 2.216 | |
| ND | 4 | |||||
| Cytogenetics groups | 0.141 | |||||
| Favorable-risk (CBF) | 54 | 22.6 | 0.0006–3.780 | 0.0137 | 0.138 ± 0.543 | |
| Intermediate-risk | 148 | 61.9 | 0.0006–12.870 | 0.0096 | 0.417 ± 1.547 | |
| Poor-risk | 37 | 15.5 | 0.0026–7.056 | 0.0170 | 0.416 ± 1.473 | |
| Flt3-ITD | 0.001 | |||||
| Flt3-ITD+ | 38 | 19.8 | 0.0006–12.870 | 0.0820 | 0.871 ± 2.275 | |
| Flt3-ITD− | 154 | 80.2 | 0.0006–7.056 | 0.0105 | 0.205 ± 0.918 | |
| NPM1 | 0.818 | |||||
| NPM1+ | 36 | 18.8 | 0.0013–12.870 | 0.0117 | 0.452 ± 2.137 | |
| NPM1− | 156 | 81.2 | 0.0006–7.056 | 0.0114 | 0.310 ± 1.058 | |
| NPM1+FLT3-ITD− | 0.015 | |||||
| NPM1+FLT3-ITD− | 22 | 11.4 | 0.0013–0.0386 | 0.0056 | 0.0104 ± 0.0102 | |
| NonNPM1+FLT3-ITD− | 170 | 88.6 | 0.0006–12.870 | 0.0128 | 0.377 ± 1.396 | |
| CEBPA | 0.034 | |||||
| CEBPADM+ | 19 | 9.9 | 0.0006–0.0848 | 0.0056 | 0.0138 ± 0.0197 | |
| CEBPADM− | 173 | 90.1 | 0.0006–12.870 | 0.0126 | 0.372 ± 1.389 | |
N: number; ND: not determined; CEBPADM: CEBPA double mutations
In order to determine high versue low IL2RA mRNA expression levels, we used a minimal p-value approach and comprehensively analyzed on correlations of IL2RA expression with clinical outcome endpoints (CR, RFS and OS). Based on the results, we set IL2RA mRNA expression cutoff value at 80 percentiles (Additional file 1: Table S2). Correlation analysis between patients’ characteristics with IL2RA mRNA expression level as dichotomous variable were then performed. There was significantly higher frequencies of high IL2RA mRNA level cases in patients with FLT3-ITD mutation (p < 0.001, Table 3) or with t(16;21);FUS-ERG (p = 0.040, Table 2) abnormality and there was significantly lower frequencies of high IL2RA mRNA level cases in patients with NPM1+FLT3-ITD− mutational status (p = 0.005, Table 2). There was no significant association between IL2RA mRNA expression level and other cytogenetic or genetic abnormalities (Table 2) or other general features of patients’ characteristics (Additional file 1: Table S3).
Table 3.
Uni and multivariable analysis on CR status, RFS and OS in AML cohort
| Variable | Univariable | Multivariable | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CR | RFS | OS | CR | RFS | OS | ||||
| p-value | p-value | p-value | p-value | HR (95% CI) | p-value | HR (95% CI) | p-value | HR (95% CI) | |
| Age group | 0.038 | 0.306 | 0.002 | 0.040 | 1.485 (1.017–2.168) | ||||
| High WBC counts | 0.880 | 0.603 | 0.929 | ||||||
| High blast percentage | 0.443 | 0.173 | 0.964 | ||||||
| Secondary AML | 0.023 | 0.243 | 0.010 | ||||||
| Cytogenetics risk groups | 0.006 | 0.001 | < 0.001 | 0.010 | 2.024 (1.188–3.449) | 0.045 | 1.400 (1.007–1.947) | < 0.001 | 2.417 (1.480–3.948) |
| High IL2RA mRNA level | 0.005 | < 0.001 | < 0.001 | 0.008 | 2.17 (1.281–5.346) | < 0.001 | 4.008 (2.437–6.592) | < 0.001 | 3.448 (1.839–6.462) |
| FLT3-ITD+ | 0.183 | 0.019 | 0.018 | ||||||
| NPM1+ | 0.963 | 0.955 | 0.507 | ||||||
| NPM1+FLT3-ITD− | 0.759 | 0.269 | 0.291 | ||||||
| c-KIT D816V+ | 0.845 | 0.022 | 0.668 | 0.015 | 3.145 (1.251–7.0901 | ||||
| CEBPADM+ | 0.998 | 0.813 | 0.061 | ||||||
Table 2.
Correlation of IL2RA mRNA expressions with cytogenetical and genetic abnormalities in AML
| Cytogenetical and genetic abnormalities | N/Total | Low IL2RA mRNA expression | High IL2RA mRNA expression | p-value |
|---|---|---|---|---|
| CBF | 54/239 | 45/191 | 9/48 | 0.565 |
| Intermediate-risk | 148/239 | 118/191 | 30/48 | 1.000 |
| Poor-risk | 39/239 | 29/191 | 9/48 | 0.505 |
| t(8;21);RUNX1-RUNX1T1 | 37/239 | 29/191 | 8/48 | 0.824 |
| inv(16) or t(16;16);CBFB-MYH11 | 17/239 | 16/191 | 1/48 | 0.207 |
| t(9;11);MLLT3-KMT2A | 4/239 | 4/191 | 0/48 | 0.586 |
| t(6;9);DEK-NUP214 | 3/239 | 2/191 | 1/48 | 0.491 |
| inv(3) or t(3;3);GATA2, MECOM | 6/239 | 4/191 | 2/48 | 0.347 |
| 11q23; non t(9;11) | 3/239 | 3/191 | 0/48 | 0.700 |
| t(16;21);FUS-ERG | 2/239 | 0/191 | 2/48 | 0.040 |
| −7/7q− | 11/239 | 7/191 | 4/48 | 0.238 |
| −5/5q− | 5/239 | 4/191 | 1/48 | 1.000 |
| Complex | 17/239 | 12/191 | 5/48 | 0.346 |
| Normal karyotype | 113/239 | 92/191 | 21/48 | 0.630 |
| Trisomy | 17/239 | 13/191 | 4/48 | 0.754 |
| FLT3-ITD+ | 38/192 | 17/151 | 21/41 | < 0.001 |
| NPM1+ | 36/192 | 28/151 | 8/41 | 1.000 |
| NPM1+FLT3-ITD− | 22/192 | 22/151 | 0/41 | 0.005 |
| CEBPADM+ | 19/192 | 18/151 | 1/41 | 0.082 |
| c-KIT D816V+ | 7/192 | 6/151 | 1/41 | 1.000 |
N: number
Correlation of high IL2RA mRNA level with poor clinical outcomes in AML cohort
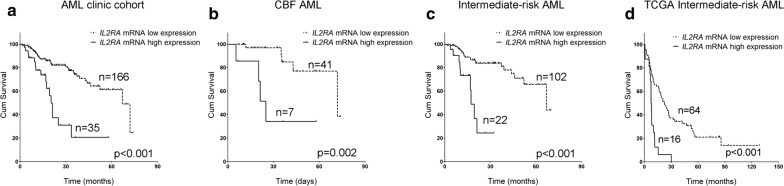
We further sought to examine if mRNA expression of IL2RA, in the context of other known important clinical or laboratory markers, correlates with clinical outcomes including CR status after two cycles of induction chemotherapy, RFS and OS in the cohort of 239 AML patients. Univariable analyses showed that high IL2RA mRNA level significantly correlated with CR status (p = 0.005) shorter RFS (p < 0.001) and shorter OS (p < 0.001, Table 3 and Fig. 2a). Other parameters, such as age, AML type (secondary vs. de novo), cytogenetics risk group, FLT3-ITD or c-KIT D816V mutation status also appeared to be significant predictors of clinical outcomes (Table 3). The prognostic value of IL2RA mRNA expression level was further confirmed by multivariable analyses, which showed that high IL2RA expression (p = 0.008, HR = 2.617) and cytogenetics risk status (p = 0.010, HR = 2.024) remained significant in predicting CR status, high IL2RA expression (p < 0.001, HR = 4.008), cytogenetics risk status (p = 0.045, HR = 1.400) and c-KIT D816V (p = 0.015, HR = 3.145) remained significant in predicting shorter RFS and high IL2RA expression (p < 0.001, HR = 3.448), cytogenetics risk group (p < 0.001, HR = 2.417) and age (p = 0.040, HR = 1.485) remained significant in predicting shorter OS (Table 3).
Fig. 2.
Survival analysis of OS by IL2RA mRNA expression levels in AML cohorts. a In our AML clinical cohort, the OS of patients with higher IL2RA expression are significantly shorter than those with lower expression levels (p < 0.001), with the median OS 20.3 months vs. 66.9 months. b In CBF AML, the OS of patients with higher IL2RA expression are significantly shorter than those with lower expression levels (p = 0.002), with the median OS 25.0 months vs. 71.8 months. c In intermediate-risk AML, the OS of patients with higher IL2RA expression are significantly shorter than those with lower expression levels (p < 0.001), with the median OS 17.3 months vs. 66.9 months. d In 80 TCGA intermediate-risk AML cases, higher IL2RA expression showed significantly shorter OS (p < 0.001), with the median OS 7.5 months vs. 22.3 months
High IL2RA mRNA expression level predicted adverse clinical outcome in core binding factor (CBF) AML and intermediate-risk AML
Within our cohort, AML of favourable-risk group only contain CBF AML with t(8;21);RUNX1-RUNX1T1 or inv(16);CBFB-MYH11 cytogenetics, since APL were not included. In the AML cohort, there were 9 out of 54 (16.7%) cases with high IL2RA expression in CBF AML subgroup, 30 out of 145 (20.3%) cases with high IL2RA expression in intermediate-risk AML and 9 out of 37 (24.3%) cases in poor-risk AML subgroup. Although high IL2RA expression cases appeared to be more frequent in AML of poor risk than in AML of favourable- or intermediate-risk, the difference was not significant (p = 0.667, Additional file 1: Table S3). We further characterized the prognostic value of IL2RA mRNA expression in CBF AML, intermediate-risk AML and poor-risk AML.
Within the CBF AML subgroup, IL2RA mRNA expression status was significantly associated with age (p = 0.026) and FLT3-ITD (p = 0.023) status (Table 4), but not with c-KIT D816V mutation (p = 1.000, Table 4). In CBF AML, univariable analyses by Kaplan–Meier method showed that high IL2RA mRNA level correlated with shorter RFS (p < 0.001) and shorter OS (p = 0.001, Table 5 and Fig. 2b). Further multivariable analyses confirmed high IL2RA mRNA level (p = 0.002, HR = 5.872), along with c-KIT D816V (p = 0.013, HR = 4.309) remained as significant in predicting shorter RFS and high IL2RA mRNA level (p = 0.014, HR = 5.718) alone remained significant in predicting shorter OS in this particular subtype of AML (Table 5).
Table 4.
Association of IL2RA mRNA expression with prognosis relevant variables in CBF, intermediate-risk and poor-risk AML
| CBF AML | Intermediate-risk AML | Poor-risk AML | |||||||
|---|---|---|---|---|---|---|---|---|---|
| High IL2RA mRNA | Low IL2RA mRNA | p-value | High IL2RA mRNA | Low IL2RA mRNA | p-value | High IL2RA mRNA | Low IL2RA mRNA | p-value | |
| Age group | 0.026 | 0.813 | 0.348 | ||||||
| 15–29 | 5 (9) | 16 (45) | 8 (30) | 26 (118) | 1 (9) | 9 (28) | |||
| 30–49 | 1 (9) | 25 (45) | 15 (30) | 59 (118) | 3 (9) | 10 (28) | |||
| 50–65 | 3 (9) | 4 (45) | 7 (30) | 33 (118) | 5 (9) | 9 (28) | |||
| High WBC counts | 2 (8) | 12 (30) | 0.686 | 9 (18) | 29 (78) | 0.423 | 1 (9) | 5 (23) | 0.648 |
| High blast percentage | 5 (9) | 32 (45) | 0.439 | 18 (30) | 72 (118) | 1.000 | 3 (9) | 19 (28) | 0.118 |
| Secondary AML | 0 (9) | 0 (45) | NA | 1/30 | 2/118 | 0.496 | 2/9 | 3 (28) | 0.577 |
| FLT3-ITD+ | 2 (7) | 0 (36) | 0.023 | 17 (27) | 14 (90) | < 0.001 | 2 (7) | 3 (25) | 0.296 |
| NPM1+ | 0 (7) | 0 (36) | NA | 8 (27) | 28 (90) | 1.000 | 0 (7) | 0 (25) | NA |
| NPM1+FLT3-ITD− | 0 (7) | 0 (36) | NA | 0 (27) | 22 (90) | 0.002 | 0 (7) | 0 (25) | NA |
| CEBPADM+ | 0 (7) | 0 (36) | NA | 1 (27) | 18 (90) | 0.070 | 0 (7) | 0 (25) | NA |
| c-Kit D816V+ | 1 (7) | 4 (36) | 1.000 | 0 (27) | 1 (90) | 1.000 | 0 (7) | 1 (25) | 1.000 |
The number in the parentheses is the number of patients that are of high or low IL2RA expression tested in that particular parameter
NA not applicable
*The p value for RFS and OS is calculated by Kaplan–Meier log-rank test. The p value for the other parameters are calculated by Fisher’s exact test or Chi-Square test
Table 5.
Uni and multivariable analyses on RFS and OS in CBF AML
| Variable | Univariable | Multivariable | ||||
|---|---|---|---|---|---|---|
| RFS | OS | RFS | OS | |||
| p value | p value | p value | HR (95% CI) | p value | HR (95% CI) | |
| Age group | 0.549 | 0.053 | ||||
| High WBC counts | 0.343 | 0.197 | ||||
| High blast percentage | 0.056 | 0.344 | ||||
| High IL2RA mRNA level | < 0.001 | 0.002 | 0.002 | 5.872 (1.902–18.129) | 0.014 | 5.718 (1.423–22.984) |
| FLT3-ITD+ | 0.526 | 0.017 | ||||
| c-Kit D816V+ | 0.006 | 0.398 | 0.013 | 4.309 (1.365–13.607) | ||
Within the intermediate-risk AML subgroup, high IL2RA mRNA expression level was significantly associated with FLT3-ITD (p < 0.001, Table 5) and NPM1+FLT3-ITD− (p = 0.002, Table 5) mutational status. High IL2RA mRNA expression significantly correlated with lower CR rate (p = 0.018, Table 5), shorter RFS (p < 0.001) and shorter OS (p < 0.001) by univariable analysis (Table 6 and Fig. 2c). As well, high IL2RA mRNA level remain as significant predictor for shorter RFS (p < 0.001, HR = 6.637) and shorter OS (p < 0.001, HR = 5.211, Table 6 and Fig. 2c) by multivariable analyses within this subgroup of AML.
Table 6.
Uni and multivariable analyses on RFS and OS in intermediate-risk AML
| Variable | Univariable | Multivariable | ||||
|---|---|---|---|---|---|---|
| RFS | OS | RFS | OS | |||
| p value | p value | p value | HR (95% CI) | p value | HR (95% CI) | |
| Age group | 0.633 | 0.223 | ||||
| High WBC counts | 0.512 | 0.300 | ||||
| High blast percentage | 0.759 | 0.779 | ||||
| Secondary AML | 0.705 | 0.168 | ||||
| High IL2RA mRNA level | < 0.001 | < 0.001 | < 0.001 | 6.637 (3.424–12.865) | < 0.001 | 5.211 (2.129–12.752) |
| FLT3-ITD+ | 0.006 | 0.006 | ||||
| CEBPADM+ | 0.930 | 0.071 | ||||
| NPM1+ | 0.685 | 0.898 | ||||
| NPM1+FLT3-ITD− | 0.440 | 0.478 | ||||
Within the poor-risk AML subgroup, however, high IL2RA mRNA expression was not associated with relevant prognostic clinical or laboratory parameters (Table 4) nor predicted worse RFS or survival within this subgroup (Additional file 1: Table S4).
Association of high IL2RA expression with known prognostic mRNA expression biomarkers in intermediate-risk AML by NanoString
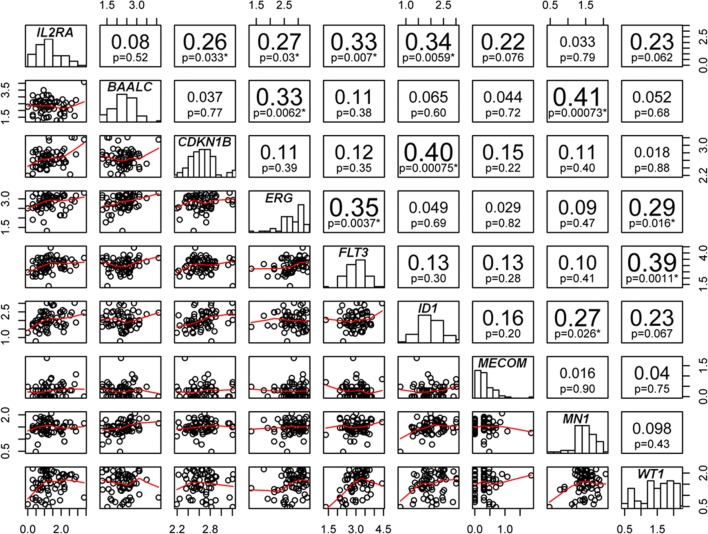
The prognostic value of many mRNA expression biomarkers was previously indicated in intermediate-risk AML and we sought to examine if IL2RA mRNA expression is correlated to these prognostic mRNA expression markers and if its prognostic value is independent of these biomarkers. We developed a NanoString multiplexed gene panel (Additional file 1: Table S5) and simultaneously measured the mRNA expressions of 8 prognostic genes (BAALC, CDKN1B, ERG, MECOM/EVI1, FLT3, ID1, MN1 and WT1) as well as IL2RA in an intermediate-risk AML cohort 66 patients. The clinical characteristics of these patients were described in Additional file 1: Table S6. The mRNA expression of IL2RA were found to significantly associate with expression of four other prognostic genes including ID1 (p = 0.006, Spearman R = 0.335, FLT3 (p = 0.007, Spearman R = 0.329), ERG (p = 0.030, Spearman R = 0.267) and CDKN1B (p = 0.033, Spearman R = 0.263) (Fig. 3) in patients with intermediate-risk AML.
Fig. 3.
Correlation analysis of nine gene expression prognostic markers within intermediate-risk AML. The mRNA expressions of the nine AML gene expression prognostic genes were quantified by NanoString technology in 66 intermediate-risk AML. The correlations between the genes were done by bivariate correlation analysis. The scatter plots, the Spearman r and p values of the correlations are displayed. Correlations with p ≤ 0.05 were labeled by *
Added prognostic value of high IL2RA mRNA expression to known mRNA expression prognostic biomarkers in independent intermediate-risk AML cohorts
In our intermediate-risk AML cohort based on NanoString assay, we further examined the prognostic value of IL2RA in the context of eight other mRNA expression biomarkers. The cutoff value for high vs. low expression of IL2RA was set at 80th percentile, for MECOM/EVI1 at 90th percentile [46] and for the rest of genes at 50th percentile [47]. Univariable analyses showed that high IL2RA, ERG, FLT3 and WT1 mRNA expression levels correlated with worse clinical outcomes including RFS and OS in intermediate-risk AML (Table 7). In particular, high IL2RA mRNA level predicted shorter RFS (p < 0.001 and Table 7) and shorter OS (p = 0.004, Table 7) in this cohort of intermediate-risk AML. Further multivariable analysis showed that high IL2RA (p < 0.001, HR = 8.171) and FLT3 (p = 0.008, HR = 3.314) mRNA expressions remained significant as predictors for shorter RFS, whereas high ERG (p = 0.008, HR = 5.541) and IL2RA (p = 0.044, HR = 2.765) mRNA expressions remained significant for shorter OS (Table 7). We also generated receiver operating characteristic (ROC) curves on the nine prognostic mRNA gene expression markers to test their performance in predicting relapse (Additional file 1: Figure S3) or survival (Additional file 1: Figure S4). The ROC curves showed superior performance of IL2RA in predicting relapse and survival than most other potential prognostic markers. In addition, the added prognostic value of IL2RA and FLT3 was observed in predicting relapse (Additional file 1: Figure S3); IL2RA and ERG in predicting survival (Additional file 1: Figure S4).
Table 7.
Uni- and multivariable analyses of multiple prognostic gene expression biomarkers in cytogenetically intermediate-risk AML
| Prognostic markers | Univariable | Multivariable | ||||
|---|---|---|---|---|---|---|
| RFS | OS | RFS | OS | |||
| p value | p value | p value | HR (95% CI) | p value | HR (95% CI) | |
| BAALC high vs. low | 0.070 | 0.927 | ||||
| CDKN1B high vs. low | 0.229 | 0.583 | ||||
| ERG high vs. low | 0.009 | 0.001 | 0.008 | 5.541 (1.566–19.602) | ||
| MECOM/EVI1 high vs. low | 0.513 | 0.409 | ||||
| FLT3 high vs. low | 0.001 | 0.006 | 0.008 | 3.314 (1.372–8.008) | ||
| ID1 high vs. low | 0.308 | 0.216 | ||||
| IL2RA high vs. low | < 0.001 | 0.004 | < 0.001 | 8.171 (2.750–24.279) | 0.044 | 2.765 (1.028–7.437) |
| MN1 high vs. low | 0.847 | 0.211 | ||||
| WT1 high vs. low | 0.002 | 0.006 | ||||
In the intermediate-risk AML subgroup (n = 80) of TCGA-LAML cohort, we also examined the prognosis value of the nine prognostic genes by uni- and multivariable analyses. In this independent study, high IL2RA mRNA expression level alone was shown to independently predict adverse outcomes including shorter EFS (p < 0.001 and HR = 3.276; Additional file 1: Table S7) and shorter OS (p < 0.001 and HR = 3.515; Fig. 2d and Additional file 1: Table S7). ROC curves on the nine mRNA gene expression markers were additionally generated and the ROC curve of IL2RA showed superior performance than other potential prognostic biomarkers in predicting clinical outcomes in the intermediate-risk AML subgroup within the TCGA-LAML cohort (Additional file 1: Figures S5, S6).
Discussion
The prognostic value of CD biomarkers has been increasingly recognized in the AML research community over the last decade to help improve the current standard prognostic tools in AML clinical practice [2, 5, 14]. These CD markers usually predict inferior clinical outcome in AML. Specifically, some of them were shown to offer additional adverse prognostic value to current stratification strategy, such as CD25 and CD56 in cytogenetically intermediate-risk AML [5, 48] and CD56 in AML with t(8;21);AML1/ETO or with t(15;17);PML/RARA [14, 49]. While the majority of these CD biomarkers were investigated at protein level using flow cytometry technology, the prognostic value of these CD biomarkers at mRNA level remains largely unknown. Our study is a first study that emphasized on prognostic value of mRNA expressions of CD marker genes in a systemic manner and we established that mRNA expression of IL2RA gene, among a selected list of CD marker genes, is a significant and independent prognostic biomarker in AML, in particular, in CBF and intermediate-risk AML subtypes.
Our study quantified IL2RA mRNA expression in 239 AML excluding APL and showed that IL2RA mRNA is differentially expressed in groups classified by FLT3-ITD, CEBPADM or NPM1+FLT3-ITD− status, the recurrent genetic mutations that are prognostic relevant. Through a minimal p-value approach, we determined the optimal cutoff value for IL2RA mRNA expression that facilitate its use as a prognostic tool. We found that high IL2RA mRNA level is positively or negatively correlated with recurrent mutational and cytogenetic aberrations including FLT3-ITD, NPM1+FLT3-ITD−, and t(16;21);FUS/ERG, which was consistent with previous reports on CD25 [3, 5]. Our results by uni- and multivariable analyses clearly showed that high IL2RA mRNA expression was correlated with lower CR rate, shorter RFS and OS in AML and the effect is independent on other prognostic factors such as age, cytogenetic, FLT3-ITD and c-KIT D816V status. Therefore, we established the significant and independent role of high IL2RA mRNA expression as an adverse prognostic factor in AML.
While IL2RA could provided additional prognostic information to cytogenetics in AML, we further studied its prognostic value in AML of distinct cytogenetics risk statuses. CBF AML is a group of AML defined by carrying transcripts t(8;21);AML1/ETO or inv(16);CBF/MYH11. Although CBF AML belongs to the favorable-risk group, it still demonstrated considerable clinical, pathophysiological and molecular heterogeneity. In CBF AML, around 85% cases achieve CR after induction therapy, about 40–50% cases relapse after CR and around 50% cases remain alive at 5 years [50, 51]. In previous studies, the prognostic value of CD25 protein expression in favorable-risk AML or CBF AML has not been reported probably due to the low frequency of CD25 positive cases in favorable-risk group in the cohorts [5, 7]. Although the frequency of high expression IL2RA cases was also low in APL (2 out of 63, Additional file 1: Table S8) and in all favourable-risk cases in our study, the frequency of high IL2RA mRNA expression cases in CBF AML (16.7%) was not significantly different from that in intermediate-risk and poor-risk AML. Further survival analyses within this subgroup of patients demonstrated strong association between high IL2RA expression and shorter RFS and OS, and it is the first time that elevated expression of CD25 biomarker gene are indicated in predicting adverse outcome in CBF AML.
Mutations in genes activating tyrosine kinase signaling (including c-KIT, N/KRAS, and FLT3) have been shown as the most frequent additional mutations in CBF AML that confer worse prognosis [51]. Of these mutations, the c-KIT D816V has been indicated to predict poor prognosis, particularly in AML with t(8;21) [51, 52]. Correlation analysis in our study in CBF AML showed no evidence of correlation between high IL2RA expression and c-KIT D816V status (p = 1.000). Further uni- and multivariable survival analysis indicateded that high IL2RA expression and c-KIT D816V mutation remained as two independent prognostic factors to predict shorter RFS, whereas high IL2RA expression alone remained significant in predicting shorter OS in CBF AML. Moreover, high IL2RA expression significantly correlated with FLT3-ITD mutation in CBF AML (p = 0.023), similar as in intermediate-risk AML. However, it should be noted that since FLT3-ITD mutation was present at a much lower frequency in CBF-AML (4.7% in our study and 7% in literature) than in intermediate-risk AML [51], the interaction between high IL2RA expression and FLT3-ITD mutation should be rather limited. It remained to be studied if the prognostic significance of high IL2RA mRNA expression in CBF AML is associated with gene mutations involved in activating tyrosine kinase signaling other than FLT3-ITD or c-KIT D816V mutation, or with other mechanisms. Nevertheless, our results suggested the significant potential of high IL2RA mRNA, in coordination with c-KIT D816V mutation, to further refine prognostification in CBF AML and warrant future validation in larger cohort of CBF-AML.
The prognostic value of IL2RA or CD25 in intermediate-AML has been consistently shown in our study and previous studies [5–7]. However, it was unknown if IL2RA prognostic value is associated with or dependent on other gene expression prognostic biomarkers which has been well studied in intermediate-risk or normal karyotype AML. In our intermediate-risk AML cohort, a panel of prognostic gene expression markers including BAALC [15], CDKN1B [15], ERG [15, 18], FLT3 [53], ID1 [17, 47], IL2RA, MN1 [15, 19], MECOM/EVI1 [46], and WT1 [54, 55] were quantified by NanoString technology. Despite the strong correlations of IL2RA expression with other mRNA biomarkers were shown, IL2RA and FLT3 mRNA expressions remained significant in predicting shorter RFS, whereas ERG and IL2RA mRNA expressions remained significant in predicting shorter OS by uni- and multivariable analyses. Independent analysis on the same panel of genes in intermediate-risk AML cases in TCGA-LAML cohort consistently demonstrated the significant and independent value of IL2RA mRNA expression in predicting inferior clinic outcome. So far, various mRNA expression gene-panels have been designed for better stratification in AML [22, 47] and our results support the incorporation of IL2RA gene into such multi-gene panel to improve prognostification within intermediate-risk AML.
The mechanisms by which IL2RA is prognostic in AML can be implicated by its correlation results with other mRNA expression biomarkers. It is first shown by our study that upregulation of IL2RA was correlated with upregulation of FLT3 tyrosine kinase transcripts [53] and of transcription factor ID1, a key common target of oncogenic tyrosine kinases that contribute to transformation of leukemias [56], indicating a strong association of IL2RA expression with tyrosine kinases pathways. Our study also showed significant correlation of IL2RA with ERG and CDKN1B, the expressions of which have been indicated in stem cell-like featured gene signatures [57, 58]. Previously, it has been proposed that CD25 is a surrogate marker for leukemia stem cell (LSC) [4, 5], our results again provided evidence that IL2RA/CD25 is involved in the crosstalk of LSC related signalling and support that IL2RA is an indicator of the LSC signature which has been shown as a fundamental adverse prognostic feature in AML [59]. In addition to these cell autonomous mechanisms, there may be non-cell autonomous mechanisms that contribute to the prognostic role of IL2RA mRNA expression such as IL2RA expression regulatory T cells [60] or IL2/IL3 interplay [61], which warrant future studies.
Conclusions
In conclusion, our findings established the significant and independent prognostic value of high IL2RA mRNA expression in AML, particularly in CBF and intermediate-risk AML, supporting the application of high IL2RA mRNA expression as a prognostic tool to improve current stratification strategies in AML.
Additional file
Additional file 1. Additional tables and figures.
Acknowledgements
Not applicable.
Abbreviations
- AML
acute myeloid leukemia
- APL
acute promyelocytic leukemia
- BM
bone marrow
- CBF
core binding factor
- CD
cluster of differentiation
- CR
complete remission
- CXCR4
C-X-C chemokine receptor type 4
- Fab
French–American–British
- FCM
flow cytometry
- NPM1
nucleophosmin 1
- FLT3-ITD
FMS-like tyrosine kinase 3-internal tandem duplication
- HCT
hematopoietic stem cell transplantation
- HR
hazard ratio
- IL2RA
interleukin 2 receptor subunit alpha
- RQ-PCR
realtime quantitative-polymerase chain reaction
- RFS
relapse-free survival
- OS
overall survival
Authors’ contributions
WD, JH, HL, SH and DH designed the study. WZ and SS performed the RQ-PCR and NanoString experiments. WD, JL, WL, YM were involved in FCM experiments and analyses of FCM results. YH and JZ performed cytogenetic analysis. XL, WZ and SS performed mutational analysis. JH, YD and CL collected the clinical data and did the follow-up. JH, LB and YD were involved in the care of patients. SL and AG performed analyses on TCGA data. JZ, XL, DanH and DH performed statistical analyses. WD, JH, SG, XF and DH wrote, reviewed and revised the manuscript. DH, WD and JH led the whole project. All authors read and approved the final manuscript.
Funding
The research was supported from Grants 81201552, 81300409 and 81072170 funded by National Natural Science Foundation of China and Grant 2018CFB485 by Hubei Provincial Natural Science Foundation of China.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study conformed to the ethical guidelines of the World Medical Association Declaration of Helsinki. Ethics approval was obtained from ethics committee of Tongji Medical College, Huazhong University of Science and Technology. Informed consent to participate was obtained from all patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Wen Du and Jing He contributed equally to this work
Contributor Information
Wen Du, Email: mtxydw@hotmail.com.
Jing He, Email: jing937@hotmail.com.
Wei Zhou, Email: zhouwei@kindstar.com.cn.
Simin Shu, Email: shusimin@kindstar.com.cn.
Juan Li, Email: 94031453@qq.com.
Wei Liu, Email: 541745354@qq.com.
Yun Deng, Email: 619341779@qq.com.
Cong Lu, Email: 404038367@qq.com.
Shengyan Lin, Email: linshengyan@hust.edu.cn.
Yaokun Ma, Email: mayaokun@kindstar.com.cn.
Yanli He, Email: heyanlicn@hust.edu.cn.
Jine Zheng, Email: enij@sina.com.
Jiang Zhu, Email: 408387973@qq.com.
Lijuan Bai, Email: dr_lijuanbai@aliyun.com.
Junxia Yao, Email: yaojunxia@yahoo.com.
Dan Hu, Email: hudan0716@hotmail.com.
Shengqing Gu, Email: shengqing.gu@gmail.com.
Huiyu Li, Email: huiyuli26@aliyun.com.
Anyuan Guo, Email: guoay@hust.edu.cn.
Shiang Huang, Email: sa2huang@hotmail.com.
Xiaolan Feng, Email: fxiaolan@ucalgary.ca.
Dong Hu, Phone: 86-2785726015, Email: dong_hu@hust.edu.cn.
References
- 1.Dohner H, Estey E, Grimwade D, Amadori S, Appelbaum FR, Buchner T, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liersch R, Muller-Tidow C, Berdel WE, Krug U. Prognostic factors for acute myeloid leukaemia in adults—biological significance and clinical use. Br J Haematol. 2014;165(1):17–38. doi: 10.1111/bjh.12750. [DOI] [PubMed] [Google Scholar]
- 3.Ikegawa S, Doki N, Kurosawa S, Yamaguchi T, Sakaguchi M, Harada K, et al. CD25 expression on residual leukemic blasts at the time of allogeneic hematopoietic stem cell transplant predicts relapse in patients with acute myeloid leukemia without complete remission. Leuk Lymphoma. 2016;57(6):1375–1381. doi: 10.3109/10428194.2015.1099644. [DOI] [PubMed] [Google Scholar]
- 4.Cerny J, Yu H, Ramanathan M, Raffel GD, Walsh WV, Fortier N, et al. Expression of CD25 independently predicts early treatment failure of acute myeloid leukaemia (AML) Br J Haematol. 2013;160(2):262–266. doi: 10.1111/bjh.12109. [DOI] [PubMed] [Google Scholar]
- 5.Gonen M, Sun Z, Figueroa ME, Patel JP, Abdel-Wahab O, Racevskis J, et al. CD25 expression status improves prognostic risk classification in AML independent of established biomarkers: ECOG phase 3 trial, E1900. Blood. 2012;120(11):2297–2306. doi: 10.1182/blood-2012-02-414425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Terwijn M, Feller N, van Rhenen A, Kelder A, Westra G, Zweegman S, et al. Interleukin-2 receptor alpha-chain (CD25) expression on leukaemic blasts is predictive for outcome and level of residual disease in AML. Eur J Cancer. 2009;45(9):1692–1699. doi: 10.1016/j.ejca.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 7.Nakase K, Kita K, Kyo T, Ueda T, Tanaka I, Katayama N. Prognostic relevance of cytokine receptor expression in acute myeloid leukemia: interleukin-2 receptor alpha-chain (CD25) expression predicts a poor prognosis. PLoS ONE. 2015;10(9):e0128998. doi: 10.1371/journal.pone.0128998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bae MH, Oh SH, Park CJ, Lee BR, Kim YJ, Cho YU, et al. VLA-4 and CXCR4 expression levels show contrasting prognostic impact (favorable and unfavorable, respectively) in acute myeloid leukemia. Ann Hematol. 2015;94(10):1631–1638. doi: 10.1007/s00277-015-2442-8. [DOI] [PubMed] [Google Scholar]
- 9.Ahn JY, Seo K, Weinberg OK, Arber DA. The prognostic value of CXCR4 in acute myeloid leukemia. Appl Immunohistochem Mol Morphol. 2013;21(1):79–84. doi: 10.1097/PAI.0b013e3182606f4d. [DOI] [PubMed] [Google Scholar]
- 10.Mannelli F, Cutini I, Gianfaldoni G, Bencini S, Scappini B, Pancani F, et al. CXCR4 expression accounts for clinical phenotype and outcome in acute myeloid leukemia. Cytom B Clin Cytom. 2014;86B:340–349. doi: 10.1002/cytob.21156. [DOI] [PubMed] [Google Scholar]
- 11.Zhu HH, Liu YR, Jiang H, Lu J, Qin YZ, Jiang Q, et al. CD34 expression on bone marrow blasts is a novel predictor of poor prognosis independent of FlT3-ITD in acute myeloid leukemia with the NPM1-mutation. Leuk Res. 2013;37(6):624–630. doi: 10.1016/j.leukres.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 12.Raspadori D, Lauria F, Ventura MA, Rondelli D, Visani G, de Vivo A, et al. Incidence and prognostic relevance of CD34 expression in acute myeloblastic leukemia: analysis of 141 cases. Leuk Res. 1997;21(7):603–607. doi: 10.1016/S0145-2126(97)00015-5. [DOI] [PubMed] [Google Scholar]
- 13.Xu S, Li X, Zhang J, Chen J. Prognostic value of CD56 in patients with acute myeloid leukemia: a meta-analysis. J Cancer Res Clin Oncol. 2015;141(10):1859–1870. doi: 10.1007/s00432-015-1977-3. [DOI] [PubMed] [Google Scholar]
- 14.Iriyama N, Hatta Y, Takeuchi J, Ogawa Y, Ohtake S, Sakura T, et al. CD56 expression is an independent prognostic factor for relapse in acute myeloid leukemia with t(8;21) Leuk Res. 2013;37(9):1021–1026. doi: 10.1016/j.leukres.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 15.Haferlach C, Kern W, Schindela S, Kohlmann A, Alpermann T, Schnittger S, et al. Gene expression of BAALC, CDKN1B, ERG, and MN1 adds independent prognostic information to cytogenetics and molecular mutations in adult acute myeloid leukemia. Genes Chromosomes Cancer. 2012;51(3):257–265. doi: 10.1002/gcc.20950. [DOI] [PubMed] [Google Scholar]
- 16.Santamaria C, Chillon MC, Garcia-Sanz R, Perez C, Caballero MD, Mateos MV, et al. BAALC is an important predictor of refractoriness to chemotherapy and poor survival in intermediate-risk acute myeloid leukemia (AML) Ann Hematol. 2010;89(5):453–458. doi: 10.1007/s00277-009-0864-x. [DOI] [PubMed] [Google Scholar]
- 17.Tang R, Hirsch P, Fava F, Lapusan S, Marzac C, Teyssandier I, et al. High Id1 expression is associated with poor prognosis in 237 patients with acute myeloid leukemia. Blood. 2009;114(14):2993–3000. doi: 10.1182/blood-2009-05-223115. [DOI] [PubMed] [Google Scholar]
- 18.Metzeler KH, Dufour A, Benthaus T, Hummel M, Sauerland MC, Heinecke A, et al. ERG expression is an independent prognostic factor and allows refined risk stratification in cytogenetically normal acute myeloid leukemia: a comprehensive analysis of ERG, MN1, and BAALC transcript levels using oligonucleotide microarrays. J Clin Oncol. 2009;27(30):5031–5038. doi: 10.1200/JCO.2008.20.5328. [DOI] [PubMed] [Google Scholar]
- 19.Heuser M, Beutel G, Krauter J, Dohner K, von Neuhoff N, Schlegelberger B, et al. High meningioma 1 (MN1) expression as a predictor for poor outcome in acute myeloid leukemia with normal cytogenetics. Blood. 2006;108(12):3898–3905. doi: 10.1182/blood-2006-04-014845. [DOI] [PubMed] [Google Scholar]
- 20.Haferlach C, Bacher U, Kohlmann A, Schindela S, Alpermann T, Kern W, et al. CDKN1B, encoding the cyclin-dependent kinase inhibitor 1B (p27), is located in the minimally deleted region of 12p abnormalities in myeloid malignancies and its low expression is a favorable prognostic marker in acute myeloid leukemia. Haematologica. 2011;96(6):829–836. doi: 10.3324/haematol.2010.035584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hinai AA, Valk PJ. Review: Aberrant EVI1 expression in acute myeloid leukaemia. Br J Haematol. 2016;172(6):870–878. doi: 10.1111/bjh.13898. [DOI] [PubMed] [Google Scholar]
- 22.Rockova V, Abbas S, Wouters BJ, Erpelinck CA, Beverloo HB, Delwel R, et al. Risk stratification of intermediate-risk acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011;118(4):1069–1076. doi: 10.1182/blood-2011-02-334748. [DOI] [PubMed] [Google Scholar]
- 23.Marcucci G, Yan P, Maharry K, Frankhouser D, Nicolet D, Metzeler KH, et al. Epigenetics meets genetics in acute myeloid leukemia: clinical impact of a novel seven-gene score. J Clin Oncol. 2014;32(6):548–556. doi: 10.1200/JCO.2013.50.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanda Y, Hamaki T, Yamamoto R, Chizuka A, Suguro M, Matsuyama T, et al. The clinical significance of CD34 expression in response to therapy of patients with acute myeloid leukemia: an overview of 2483 patients from 22 studies. Cancer. 2000;88(11):2529–2533. doi: 10.1002/1097-0142(20000601)88:11<2529::AID-CNCR14>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 25.Junca J, Garcia O, Garcia-Caro M, Vila J, Zamora L, Cabezon M, et al. CD34 expression and the outcome of nucleophosmin 1-mutated acute myeloid leukemia. Ann Hematol. 2016;95(12):1949–1954. doi: 10.1007/s00277-016-2811-y. [DOI] [PubMed] [Google Scholar]
- 26.Dang H, Chen Y, Kamel-Reid S, Brandwein J, Chang H. CD34 expression predicts an adverse outcome in patients with NPM1-positive acute myeloid leukemia. Hum Pathol. 2013;44(10):2038–2046. doi: 10.1016/j.humpath.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 28.Battle A, Khan Z, Wang SH, Mitrano A, Ford MJ, Pritchard JK, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347(6222):664–667. doi: 10.1126/science.1260793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grun D, Kirchner M, Thierfelder N, Stoeckius M, Selbach M, Rajewsky N. Conservation of mRNA and protein expression during development of C. elegans. Cell Rep. 2014;6(3):565–577. doi: 10.1016/j.celrep.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 31.International Standing Committee on Human Cytogenetic Nomenclature. Shaffer LG, Slovak ML, Campbell LJ. ISCN 2009: an international system for human cytogenetic nomenclature. Basel: Karger; 2009. p. 2009. [Google Scholar]
- 32.Zheng J, Wang X, Hu Y, Yang J, Liu J, He Y, et al. A correlation study of immunophenotypic, cytogenetic, and clinical features of 180 AML patients in China. Cytom B Clin Cytom. 2008;74(1):25–29. doi: 10.1002/cyto.b.20368. [DOI] [PubMed] [Google Scholar]
- 33.Quentmeier H, Reinhardt J, Zaborski M, Drexler HG. FLT3 mutations in acute myeloid leukemia cell lines. Leukemia. 2003;17(1):120–124. doi: 10.1038/sj.leu.2402740. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki T, Kiyoi H, Ozeki K, Tomita A, Yamaji S, Suzuki R, et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106(8):2854–2861. doi: 10.1182/blood-2005-04-1733. [DOI] [PubMed] [Google Scholar]
- 35.Ho PA, Alonzo TA, Gerbing RB, Pollard J, Stirewalt DL, Hurwitz C, et al. Prevalence and prognostic implications of CEBPA mutations in pediatric acute myeloid leukemia (AML): a report from the Children’s Oncology Group. Blood. 2009;113(26):6558–6566. doi: 10.1182/blood-2008-10-184747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SH, Chi HS, Min SK, Park BG, Jang S, Park CJ. Prognostic impact of c-KIT mutations in core binding factor acute myeloid leukemia. Leuk Res. 2011;35(10):1376–1383. doi: 10.1016/j.leukres.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Hu D, Zhou W, Wang F, Shu SM, Fan LL, He J, et al. Development of a NanoString assay to detect leukemogenic fusion transcripts in acute myeloid leukemia. Int J Lab Hematol. 2016;38(6):663–673. doi: 10.1111/ijlh.12555. [DOI] [PubMed] [Google Scholar]
- 38.Patterson BK, Landay A, Andersson J, Brown C, Behbahani H, Jiyamapa D, et al. Repertoire of chemokine receptor expression in the female genital tract: implications for human immunodeficiency virus transmission. Am J Pathol. 1998;153(2):481–490. doi: 10.1016/S0002-9440(10)65591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beillard E, Pallisgaard N, van der Velden VH, Bi W, Dee R, van der Schoot E, et al. Evaluation of candidate control genes for diagnosis and residual disease detection in leukemic patients using ‘real-time’ quantitative reverse-transcriptase polymerase chain reaction (RQ-PCR)—a Europe against cancer program. Leukemia. 2003;17(12):2474–2486. doi: 10.1038/sj.leu.2403136. [DOI] [PubMed] [Google Scholar]
- 40.Baldus CD, Tanner SM, Ruppert AS, Whitman SP, Archer KJ, Marcucci G, et al. BAALC expression predicts clinical outcome of de novo acute myeloid leukemia patients with normal cytogenetics: a Cancer and Leukemia Group B Study. Blood. 2003;102(5):1613–1618. doi: 10.1182/blood-2003-02-0359. [DOI] [PubMed] [Google Scholar]
- 41.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, et al. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115(3):453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez P, Solenthaler M, Spertini O, Quarroz S, Rovo A, Lovey PY, et al. Using digital RNA counting and flow cytometry to compare mRNA with protein expression in acute leukemias. PLoS ONE. 2012;7(11):e49010. doi: 10.1371/journal.pone.0049010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kern W, Kohlmann A, Wuchter C, Schnittger S, Schoch C, Mergenthaler S, et al. Correlation of protein expression and gene expression in acute leukemia. Cytom B Clin Cytom. 2003;55(1):29–36. doi: 10.1002/cyto.b.10025. [DOI] [PubMed] [Google Scholar]
- 46.Groschel S, Lugthart S, Schlenk RF, Valk PJ, Eiwen K, Goudswaard C, et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J Clin Oncol. 2010;28(12):2101–2107. doi: 10.1200/JCO.2009.26.0646. [DOI] [PubMed] [Google Scholar]
- 47.Damm F, Heuser M, Morgan M, Wagner K, Gorlich K, Grosshennig A, et al. Integrative prognostic risk score in acute myeloid leukemia with normal karyotype. Blood. 2011;117(17):4561–4568. doi: 10.1182/blood-2010-08-303479. [DOI] [PubMed] [Google Scholar]
- 48.Coelho-Silva JL, Carvalho LE, Oliveira MM, Franca-Neto PL, Andrade AT, Lima AS, et al. Prognostic importance of CD56 expression in intermediate risk acute myeloid leukaemia. Br J Haematol. 2017;176(3):498–501. doi: 10.1111/bjh.13971. [DOI] [PubMed] [Google Scholar]
- 49.Breccia M, De Propris MS, Minotti C, Stefanizzi C, Raponi S, Colafigli G, et al. Aberrant phenotypic expression of CD15 and CD56 identifies poor prognostic acute promyelocytic leukemia patients. Leuk Res. 2014;38(2):194–197. doi: 10.1016/j.leukres.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 50.Sood R, Hansen NF, Donovan FX, Carrington B, Bucci D, Maskeri B, et al. Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia. Leukemia. 2016;30(2):501–504. doi: 10.1038/leu.2015.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duployez N, Marceau-Renaut A, Boissel N, Petit A, Bucci M, Geffroy S, et al. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood. 2016;127(20):2451–2459. doi: 10.1182/blood-2015-12-688705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen W, Xie H, Wang H, Chen L, Sun Y, Chen Z, et al. Prognostic Significance of KIT Mutations in Core-Binding Factor Acute Myeloid Leukemia: A Systematic Review and Meta-Analysis. PLoS One. 2016;11(1):e0146614. doi: 10.1371/journal.pone.0146614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozeki K, Kiyoi H, Hirose Y, Iwai M, Ninomiya M, Kodera Y, et al. Biologic and clinical significance of the FLT3 transcript level in acute myeloid leukemia. Blood. 2004;103(5):1901–8. doi: 10.1182/blood-2003-06-1845. [DOI] [PubMed] [Google Scholar]
- 54.Niavarani A, Herold T, Reyal Y, Sauerland MC, Buchner T, Hiddemann W, et al. A 4-gene expression score associated with high levels of Wilms Tumor-1 (WT1) expression is an adverse prognostic factor in acute myeloid leukaemia. Br J Haematol. 2016;172(3):401–11. doi: 10.1111/bjh.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Minetto P, Guolo F, Clavio M, De Astis E, Colombo N, Grasso R, et al. Combined assessment of WT1 and BAALC gene expression at diagnosis may improve leukemia-free survival prediction in patients with myelodysplastic syndromes. Leuk Res. 2015;39(8):866–73. doi: 10.1016/j.leukres.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 56.Tam WF, Gu TL, Chen J, Lee BH, Bullinger L, Frohling S, et al. Id1 is a common downstream target of oncogenic tyrosine kinases in leukemic cells. Blood. 2008;112(5):1981–92. doi: 10.1182/blood-2007-07-103010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Metzeler KH, Maharry K, Kohlschmidt J, Volinia S, Mrozek K, Becker H, et al. A stem cell-like gene expression signature associates with inferior outcomes and a distinct microRNA expression profile in adults with primary cytogenetically normal acute myeloid leukemia. Leukemia. 2013;27(10):2023–31. doi: 10.1038/leu.2013.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Seet CS, Sun C, Li J, You D, Volk A, et al. p27kip1 maintains a subset of leukemia stem cells in the quiescent state in murine MLL-leukemia. Mol Oncol. 2013;7(6):1069–82. doi: 10.1016/j.molonc.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varn FS, Andrews EH, Cheng C. Systematic analysis of hematopoietic gene expression profiles for prognostic prediction in acute myeloid leukemia. Sci Rep. 2015;24(5):16987. doi: 10.1038/srep16987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ustun C, Miller JS, Munn DH, Weisdorf DJ, Blazar BR. Regulatory T cells in acute myelogenous leukemia: is it time for immunomodulation? Blood. 2011;118(19):5084–95. doi: 10.1182/blood-2011-07-365817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nakase K, Kita K, Katayama N. IL-2/IL-3 interplay mediates growth of CD25 positive acute myeloid leukemia cells. Med Hypotheses. 2018;115:5–7. doi: 10.1016/j.mehy.2018.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional tables and figures.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.