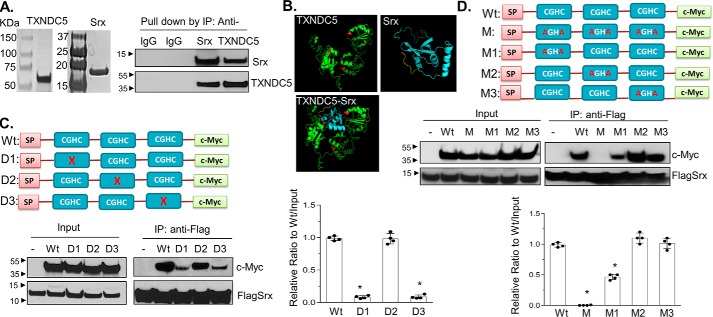
Figure 4.
Characterization and mapping of domains in TXNDC5 that directly interact with Srx. A, recombinant human Srx and TXNDC5 were purified from E. coli and visualized by Coomassie Blue staining (left). The purified proteins were mixed in the binding solution. Reciprocal IP was performed and examined by Western blotting (right). B, predicted interaction of Srx and TXNDC5 based on their structures using I-TASSER and ZDOCK software. The thioredoxin domains (with the key CGHC motif) in TXNDC5 are highlighted in red. The prediction indicates that Srx (blue) interacts with TXNDC5 (green) through the first and the third thioredoxin domain of TXNDC5. C, plasmids that express c-Myc–tagged TXNDC5 or its deletion mutants were expressed in HEK293T-FLAGSrx cells, and cell lysates were used in anti-FLAG IP and examined by Western blotting. Deletion of the first or the third CGHC motif in TXNDC5 leads to significant loss of binding to Srx. D, plasmids that express c-Myc–tagged TXNDC5 or its cysteine-specific mutants were expressed in HEK293T-FLAGSrx cells, and cell lysates were used for anti-FLAG IP and examined by Western blotting. Mutation of two cysteines in the first or the third CGHC motif in TXNDC5 leads to significant loss of binding to Srx. The bar graph with a dot plot on the right indicates the quantitative results (compared with WT; *, p < 0.05, t test). Error bars, S.D.