Polycythemia vera (PV) and essential thrombocythemia (ET) are chronic myeloproliferative neoplasms (MPNs) with high risk of thromboembolism and tendency to transform to myelofibrosis and acute leukemia. Pegylated interferon-α (PegINFa) is an effective treatment and unlike hydroxyurea (HU), it is reported to improve cytogenetic abnormalities, decrease JAK2V617F and CALR allelic burdens and achieve molecular remission [1–3]. We reported a return to polyclonal hematopoiesis in some females after PegINFa therapy [3], and unlike HU, PegINFa promoted proliferation and differentiation of PV and ET hematopoietic stem cells in vivo [4]. Cytokines, such as TNFα and interleukins are found to be elevated in MPN [5, 6] and are implicated as drivers of clonal expansion of JAK2V617F bearing cells [7]. Genome wide association studies have shown IL28 SNP and treatment with PegINFa were associated with sustained virologic response in hepatitis C [8] and suggested favorable hematological response in 20 PV/ET patients [9].
Given these observations, we analyzed the effect of PegINFa and HU treatment on molecular response, clonal hematopoiesis (females), TNFα transcript levels, and IL28B haplotype as correlated with PegINFa response. We included 82 patients diagnosed with high-risk PV (n = 45) and ET (n = 37) (per WHO 2008) treated from 2008 to March 2017. Mutational analysis for JAK2V617F and cMPL was performed by quantitative allele-specific-PCR [10], and CALR mutations by semi-quantitative fragment analysis [11]. In females, SNPs of 5 X-chromosome genes (G6PD, MPP1, FHL1, BTK and IDS) were genotyped and in heterozygotes transcriptional allelic frequencies of granulocytes were determined by quantitative allele-specific PCR. TaqMan Gene Expression Assays were used to determine TNFα and cytokine mRNAs and plasma levels of cytokines were measured. Relative TNFα expressions and JAK2V617F allelic burden were compared using paired t-test in GraphPad Prism (La Jolla, CA). As many of these patients participated in the MPD-RC trials the hematologic and clinical responses of these patients will be reported by the MPD-RC group.
JAK2V617F mutation was detectable in 70 patients, CALR mutation in 11 ET, cMPL G1544T in one ET (Supplementary Table 1). In total 31 patients (27 PV and 4 ET) in the PegINFa group, and 19 patients (17 PV and 2 ET) in the HU group had JAK2V617F mutant allelic burden >10% at baseline and with at least one 6-month follow-up allelic burden value (Supplementary Figure 1). Overall molecular response rate was 14 (45%) in PegINFa group and 17(77.2%) in the HU group. No complete molecular response rates were noted during the entire follow-up. Among 48 PegINFa treated patients, 29 had prior HU, and in this subset of HU refractory/intolerant patients, only 3 (16.6%) had molecular response. In all patients whose JAK2V617F allelic burden decreased to <5%, the allelic burden was retested using quantitative digital PCR, and their low detectable JAK2 mutant burden was confirmed (range 0.42–4.3%).
In 20 patients, JAK2V617F allele burden was measured simultaneously in marrow CD34+ and peripheral blood granulocytes, and the allelic burden in CD34+ was lower than in granulocytes (Supplementary Figure 2), consistent with previous reports [4, 12].
We were able to determine clonality in 26 females (18 PV and 8 ET; 20 on PegINF and 6 on HU), and all were clonal prior to treatment. At the end of follow-up, all 6 patients on HU continued to be clonal but 2 (10%) patients on PegINFa converted to polyclonal. Their median time of HU and PegINFa treatment was 12 months (1–70) and 30 months (5–75). IL28 SNP rs12979860 was determined in 44 patients who received PegINFa, and we found no correlation with clinical response (RR 0.86 (95%CI0.65–1.25)).
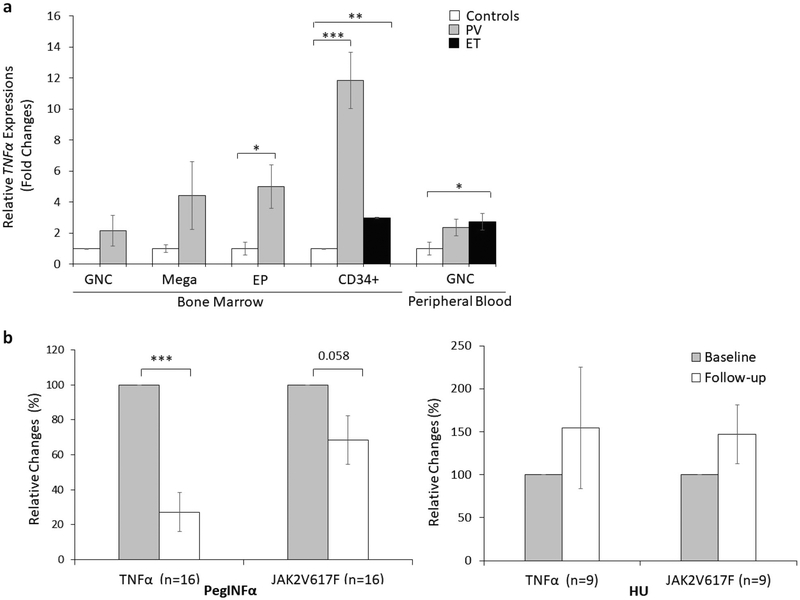
Pro-inflammatory cytokines suppress normal hematopoiesis and are elevated in MPN [5, 6]. In our initial pilot analyses, we noted increased TNFα by immunostaining in PV marrow with positive signal preferentially localized in immature cells and megakaryocytes (Supplementary Figure 3). We isolated cells of different degrees of differentiation from PV marrow and analyzed their TNFα transcript. Transcripts were higher in erythroid progenitors and the highest in CD34+ stem cells, suggesting a possible role of TNFα on suppression of PV/ET clones (Fig. 1a). We then determined TNFα transcripts in CD34 + cells on 25 patients on whom samples were available before and after treatment (PegINFa=16 and HU=9), and peripheral granulocytes on 21 patients (PegINFa=12 and HU=9). After PegINFa therapy, there was a significant decrease in TNFα expression in CD34+ cells (p = 0.0004), which paralleled the decline in JAK2V617F allele burden (Fig. 1b). The decline of both JAK2V617F allele and TNFα closely corresponded to PegINFa therapy, as shown in a patient in whom PegINFa had to be discontinued and switched to HU (Supplementary Figure 4). In HU-treated patients, there was no such decrease in either TNFα expression or JAK2V617F allelic burden (Fig. 1b). In contrast, TNFα expression in peripheral granulocytes did not decrease after either therapy and did not correlate with the JAK2V617F allelic burden. We also measured transcripts of inflammatory cytokines IL6, IL8, IL1B, and IL12A in available residual RNA. Peripheral granulocytes of PV and ET patients had higher expression of these inflammatory markers compared to controls, but did not change after PegINFa therapy in marrow CD34+ cells or granulocytes. However, after HU therapy, there was increased expression of IL8 both in marrow CD34+ cells and peripheral granulocytes, as well as increased IL12A in peripheral granulocytes (Supplementary Figure 5a–b–c). Concomitant plasma samples were available in 7 patients, which showed decreasing trend after PegINFa therapy (p = 0.056), while plasma IL6 levels (p = 0.233) and IL1B levels were unchanged (Supplementary Figure 5d). Thus, while other inflammatory markers are over-expressed in PV and ET, only TNFα shows decrease in response to PegINFa therapy.
Fig. 1.
Relative TNFα expressions and JAK2V617F allelic burden in CD34 + cells and peripheral blood granulocytes. a TNFα expressions in CD34 + cells (n = 4) and peripheral granulocytes (n = 7) were normalized against HPRT and GUSB, calculated by ΔΔCt method, and expressed as a proportion change from the average of controls taken as1. b TNFα expression levels and JAK2V617F allelic burdens before and after PegINFa (n = 16) and HU (n = 9) in marrow CD34+ cells. TNFα expression levels were normalized against HPRT and expressed as a relative fold change against baseline as 100%. Relative allelic burden of JAK2V617F was calculated against baseline as 100%. *p <0.05; **p < 0.01; ***p < 0.0001
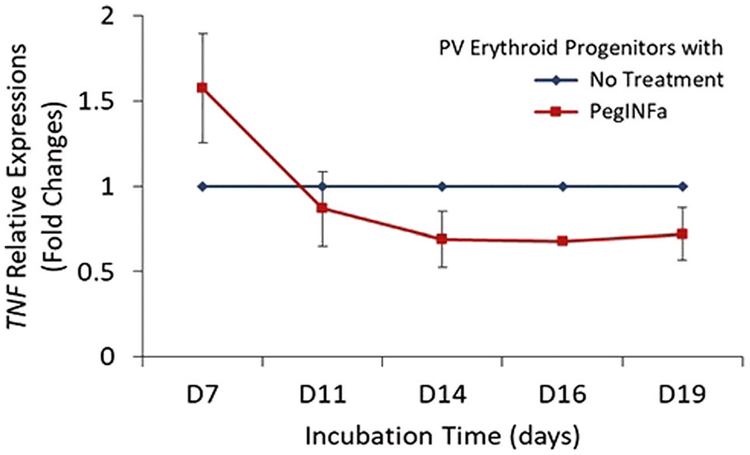
To test whether PegINFa may directly regulate TNFα in MPN cells, we cultured PV erythroid progenitors in a liquid culture system that is optimized to support the proliferation of erythroid progenitors [13, 14]. PegINFa decreased TNFα mRNA in liquid culture, mimicking the pattern seen in patients (Fig. 2). To assess the causality of TNFα on suppression of PV clone, we used TNFα inhibitor, adalimumab, in semisolid (BFU-E) and liquid erythroid cultures in 5 PV patient samples (Supplementary Figure 6a). There was initial increase of JAK2V617F allelic burden until day 7 corresponding to erythroid expansion in the culture. However, following day 11 progressive decline of JAK2V617F allelic burden is noted due to the expansion of dormant non-PV erythroid progenitors. The JAK2V617F allelic burden decreased with adalimumab by 69% on day 11 (p = 0.0047) and 49% on day 14 (p = 0.0235). Thus, the addition of adalimumab favored the outgrowth of JAK2 wild-type over JAK2V617F mutant alleles in erythroid progenitors as seen in BFU-E colonies (Supplementary Figure 6b), without any change in colony type or number of BFU-E colonies.
Fig. 2.
Effect of PegIFNa on TNF mRNA expression in erythroid progenitor cell in in vitro liquid cultures from PV patients (n = 3). Relative TNFα transcript with PegINFa was measured in erythroid progenitors and calculated compared to PV without PegIFNa as described in Fig. 1
Although PegINFa treated patients had decreased CD34+ cell TNFα transcripts which paralleled decreased JAK2V617F allelic burden, the plasma TNFα levels did not decrease to the same extent (Supplementary Figure 7). This quantitative discrepancy between TNFα mRNA expression level and its circulating protein after PegINFa therapy may suggest a paracrine inhibitory effect of TNFα. Although limited by the small number of samples, our data suggest that PegINFa suppresses TNFα expression, leading to a decrease in the JAK2 mutant clone. We did not see any change in expression of other cytokines after treatment with either PegINFa or HU, suggesting only TNFα responded to PegINFa therapy. TNFα has been shown to induce apoptosis and necroptosis in hematopoietic stem cells [15]. Thus, it is tempting to speculate that TNFα-expressing JAK2V617F clones suppress surrounding normal stem cell cells, and PegINFa abrogates this suppression, thereby permitting normal polyclonal hematopoiesis to resume.
In summary, although both PegINFa and HU decreased JAK2V617F allele burden, no complete molecular remission was achieved. However, PegINFa may restore polyclonal hematopoiesis through abrogation of TNFα expression in the bone marrow progenitor cells, suggesting possible efficacy of combination of TNF inhibitor and PegINFa in elimination of the MPN clone.
Supplementary Material
Acknowledgements
We are grateful to Dr. Bruce Beutler for advice to use TNFα inhibitor, adalimumab.
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1038/s41375-018-0080-6) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflict of interest.
References
- 1.Kiladjian JJ, Cassinat B, Chevret S, Turlure P, Cambier N, Roussel M, et al. Pegylated interferon-alfa-2a induces complete hematologic and molecular responses with low toxicity in polycythemia vera. Blood. 2008;112:3065–72. [DOI] [PubMed] [Google Scholar]
- 2.Quintas-Cardama A, Kantarjian H, Manshouri T, Luthra R, Estrov Z, Pierce S, et al. Pegylated interferon alfa-2a yields high rates of hematologic and molecular response in patients with advanced essential thrombocythemia and polycythemia vera. J Clin Oncol. 2009;27:5418–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu E, Jelinek J, Pastore YD, Guan Y, Prchal JF, Prchal JT. Discrimination of polycythemias and thrombocytoses by novel, simple, accurate clonality assays and comparison with PRV-1 expression and BFU-E response to erythropoietin. Blood. 2003;101:3294–301. [DOI] [PubMed] [Google Scholar]
- 4.King KY, Matatall KA, Shen CC, Goodell MA, Swierczek SI, Prchal JT. Comparative long-term effects of interferon alpha and hydroxyurea on human hematopoietic progenitor cells. Exp Hematol. 2015;43:912–8 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pourcelot E, Trocme C, Mondet J, Bailly S, Toussaint B, Mossuz P. Cytokine profiles in polycythemia vera and essential thrombocythemia patients: clinical implications. Exp Hematol. 2014;42:360–8. [DOI] [PubMed] [Google Scholar]
- 6.Vaidya R, Gangat N, Jimma T, Finke CM, Lasho TL, Pardanani A, et al. Plasma cytokines in polycythemia vera: phenotypic correlates, prognostic relevance, and comparison with myelofibrosis. Am J Hematol. 2012;87:1003–5. [DOI] [PubMed] [Google Scholar]
- 7.Fleischman AG, Aichberger KJ, Luty SB, Bumm TG, Petersen CL, Doratotaj S, et al. TNFalpha facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood. 2011;118:6392–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayes CN, Imamura M, Aikata H, Chayama K. Genetics of IL28B and HCV--response to infection and treatment. Nat Rev Gastroenterol Hepatol. 2012;9:406–17. [DOI] [PubMed] [Google Scholar]
- 9.Lindgren M, Pettersson H, Westin J, Lindh M, Johansson P, Andreasson B. Influence of interferon-α treatment outcome in polycythemia vera and essential thrombocythemia by genetic polymorphism in IL28B. J Hematol Malig. 2012;2:18–25. [Google Scholar]
- 10.Nussenzveig RH, Swierczek SI, Jelinek J, Gaikwad A, Liu E, Verstovsek S, et al. Polycythemia vera is not initiated by JAK2V617F mutation. Exp Hematol. 2007;35:32–8. [DOI] [PubMed] [Google Scholar]
- 11.Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369:2379–90. [DOI] [PubMed] [Google Scholar]
- 12.Moliterno AR, Williams DM, Rogers O, Isaacs MA, Spivak JL. Phenotypic variability within the JAK2 V617F-positive MPD: roles of progenitor cell and neutrophil allele burdens. Exp Hematol. 2008;36:1480–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruchova H, Yoon D, Agarwal AM, Swierczek S, Prchal JT. Erythropoiesis in polycythemia vera is hyper-proliferative and has accelerated maturation. Blood Cells Mol Dis. 2009;43:81–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaikwad A, Nussenzveig R, Liu E, Gottshalk S, Chang K, Prchal JT. In vitro expansion of erythroid progenitors from polycythemia vera patients leads to decrease in JAK2 V617F allele. Exp Hematol. 2007;35:587–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Zou Z, Wu Z, Zhao Z, Luo X, Xie C, et al. TNF-alpha-induced programmed cell death in the pathogenesis of acquired aplastic anemia. Expert Rev Hematol. 2015;8:515–26. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.