Fig. 3 |. The challenges of compound structure representation in machine learning models.
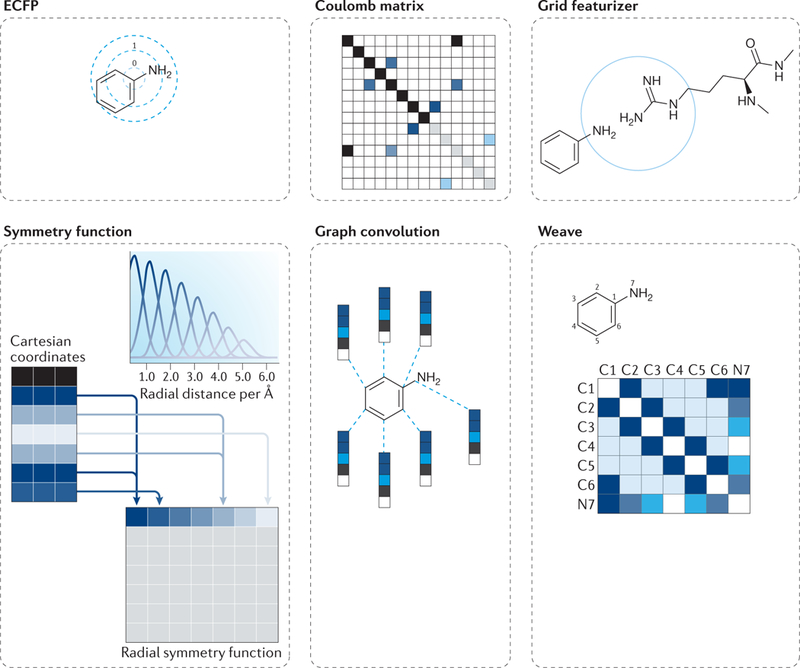
The appropriate representation of chemical structures and their features can take on many representations depending on the required application. Extended-connectivity fingerprints (ECFPs) contain information about topological characteristics of the molecule, which enables this information to be applied to tasks such as similarity searching and activity prediction. A Coulomb matrix encodes information about the nuclear charges of a molecule and their coordinates. The grid featurizer method incorporates structural features of both the ligand and the target protein as well as the intermolecular forces that contribute to binding affinity. Symmetry function is another common encoding of atomic coordinate information, which focuses on the distance between atom pairs and the on angles formed within triplets of atoms. The graph convolution method computes an initial feature vector and a neighbour list for each atom that summarizes the local chemical environment of an atom, including atom types, hybridization types and valence structures. Weave featurization calculates a feature vector for each pair of atoms in the molecule, including bond properties (if directly connected), graph distance and ring info, forming a feature matrix. Reproduced by permission of the Royal Society of Chemistry, Wu, Z. et al. MoleculeNet: a benchmark for molecular machine learning. Chem. Sci. 9, 513–530 (2018), REF.43.
