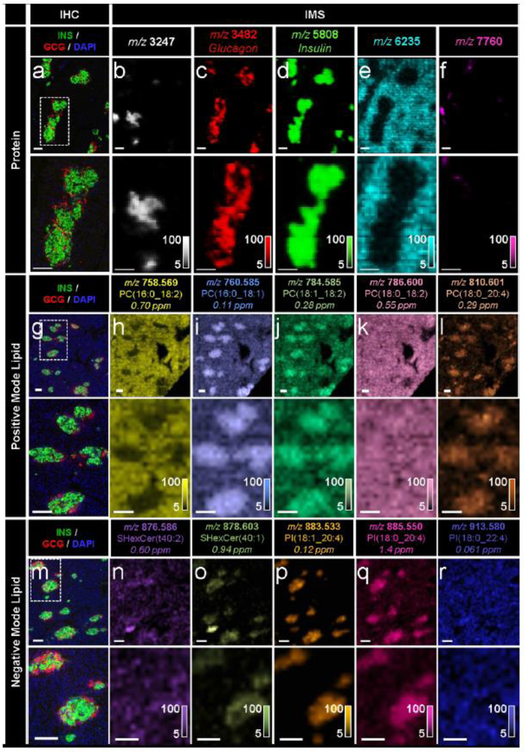
Fig. 2.
IMS enabled the untargeted detection of a wide array of molecular compounds in pancreatic tissue sections from a 19-month-old human donor, (a-f) Immunostaining (green, C- peptide; red, glucagon; blue, DAPI) and false-colour 15 μm spatial resolution IMS from serial tissue sections showing the spatial distribution of a series of proteins. Note that identification of the unknown proteins shown in (b, e, f) would require subsequent LC-MS/MS experiments. These masses were simply selected to highlight the spatial molecular diversity detectable using MALDI IMS technology, (g–l) Immunostaining and false-colour 20 μm spatial resolution IMS from serial tissue sections showing the spatial distribution of a series of PC ions, (m–r) Immunostaining and false-colour 20 μm spatial resolution IMS from serial tissue sections showing the spatial distribution of a series of sulfatide and PI ions. Lipids were identified by a combination of accurate mass measurements (ppm error reported) and MS/MS (ESM Fig. 2). Ion images are shown without normalisation and with pixel interpolation. Magnifications of the areas highlighted with white dotted lines in the IHC images are shown below each IHC and IMS image. Intensity scales of the false-colour IMS images are shown in the bottom right of each magnified image. All scale bars are 100 μm. C-PEP, C-peptide; GCG, glucagon; SHexCer, sulfatide (sulfo-hexosyl-ceramide)