FIG. 3.
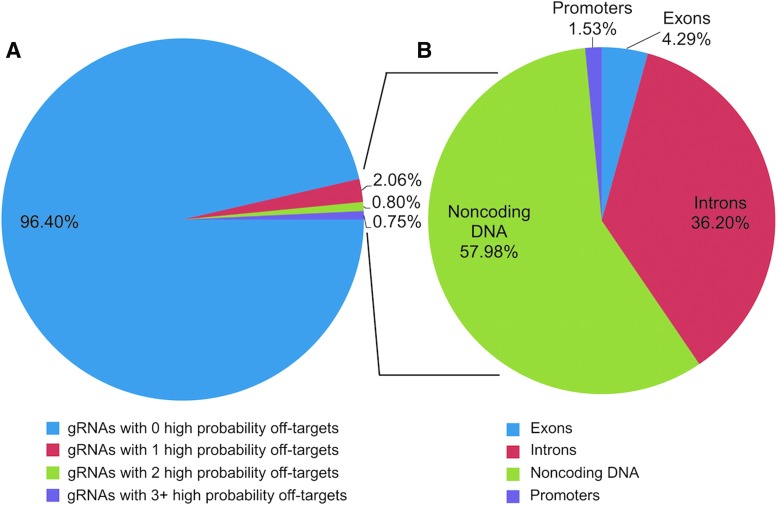
The majority (96.40%) of tested gRNA templates do not cause any off-target effects, and very few exonic off-target effects exist. (A) The total number of gRNAs with predicted off-target cleavage was calculated to be 149 of the 4,114 viable gRNAs that were created, which corresponds to 3.60% of the tested gRNAs. Eighty-five of the gRNAs only have one off-target effect (2.06% of all gRNAs), 33 (0.80%) of the gRNAs have two off-target effects, and 31 of the gRNAs have three or more off-target effects (range 3–24; median = 4). (B) These 149 gRNAs generate 326 off-target effects. Fourteen off-target effects targeted exons (4.29% of all off-targets), 118 (36.20%) targeted introns, and 189 (57.98%) targeted noncoding DNA. Eight of them generated off-target effects in promoter regions, but three of the eight hit within exons or introns. Within the pie chart, those were counted in either exonic or intronic off-target effects, so 5 (1.53%) hit within non-coding DNA.