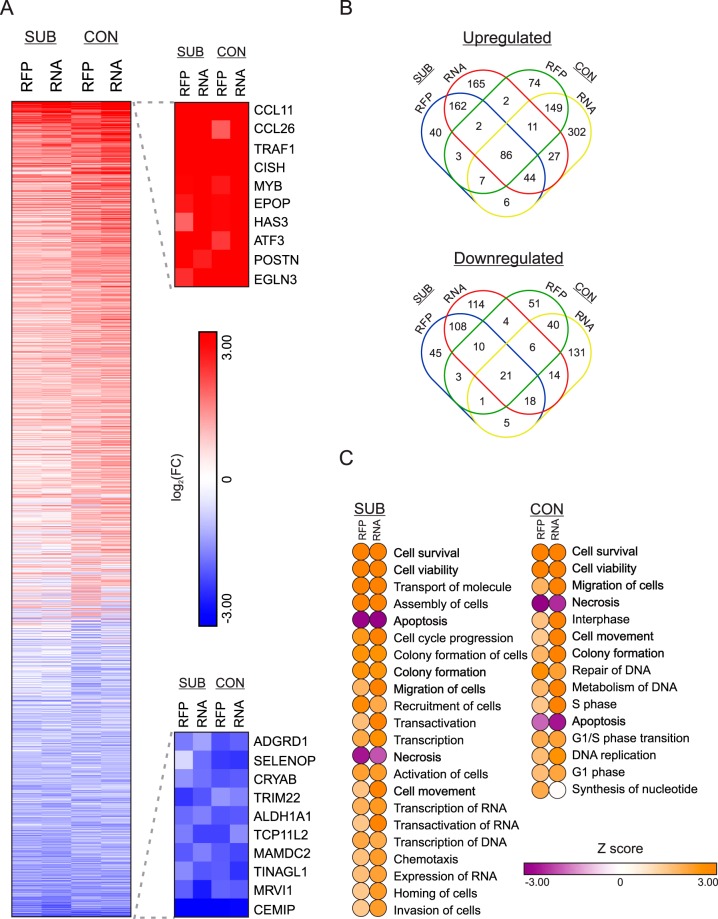
FIG 4.
Differentially regulated genes in HFF host cells following 24 h of tachyzoite infection. (A) Heat map of all genes identified in ribosome profiling (RFP) and RNA-seq (RNA) data sets. Lanes are segregated by subconfluent (SUB) and confluent (CON) sequencing libraries. Data represent log2-transformed fold change upon infection. Rows are ordered from most induced (red) to most repressed (blue). The top 10 genes from each extreme are presented. (B) Venn diagrams showing the relationship between shared and growth condition-specific up- and downregulated genes upon infection. (C) Ingenuity Pathway Analysis of modulated cellular and molecular functions in subconfluent and confluent HFFs upon tachyzoite infection. Pathways with a significance of a P value of ≤0.05 and that displayed a Z score ±2 in either the RFP or RNA data sets are shown. Pathways that are common between confluent and subconfluent HFFs are in bold.