Figure 3. An Asp (D) –to-Glu (E) “switch” in the enzymatic hinge backend underlies paralog selectivity of GSK3α and GSK3β inhibitors.
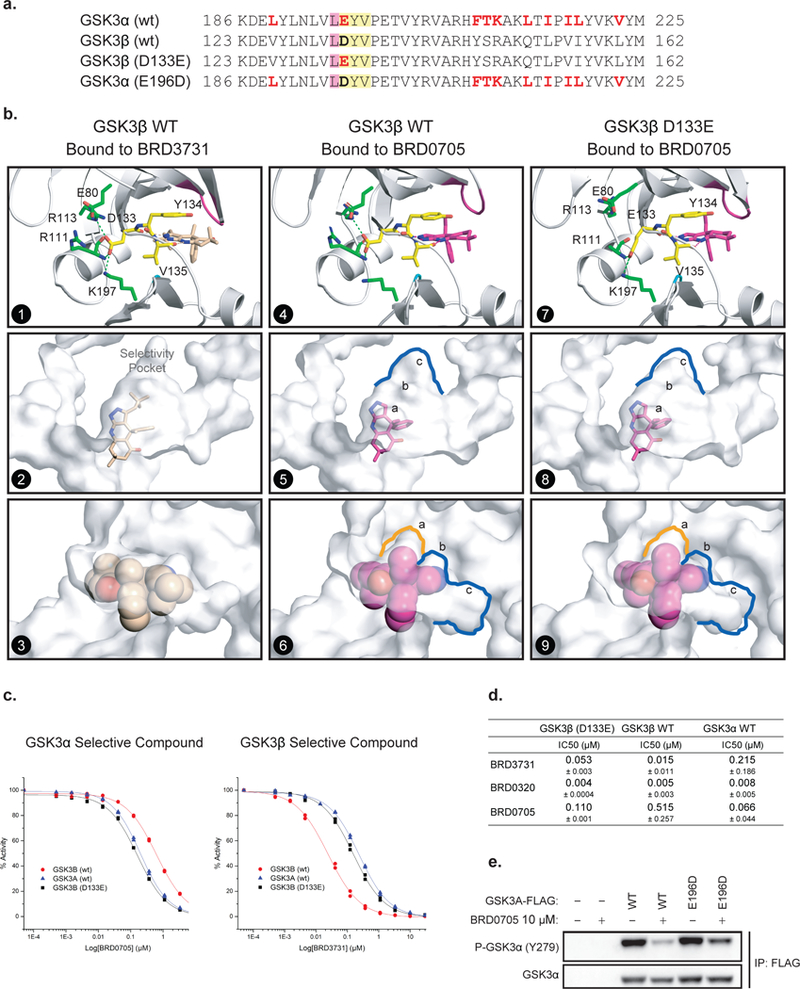
(a) Primary sequence comparison of GSK3α, GSK3β and GSK3β (D133E) and GSK3α (E196D) mutants. (b) X-ray structures of hGSK3β bound to BRD3731 (panels 1–3) and BRD0705 (panels 4–6) and hGSK3β (D133E) bound to BRD0705 (panels 7–9). The side chain of Asp133 in GSK3β forms a complex hydrogen bond network at the back of the kinase hinge and controls the shape and size of the adjacent selectivity pocket. (c-d) GSK3β (D133E) mutation “switches” compound selectivity: IC50 values for the inhibition GSK3β (D133E) were determined at Km of ATP in a motility-shift microfluidic assay (Caliper, MA) measuring phosphorylation of a synthetic substrate. Representative IC50 curves comparison in enzymatic assay using GSK3α (WT, blue), GSK3β (WT, red) and GSK3β (D133E, black) protein constructs (c). Values are the average of three or more experiments. Data are shown as IC50 values in μM ± standard deviation (d). Compounds were tested in duplicate in a 12-point dose curve with 3-fold dilution starting at 33.3 μM. (e) FLAG tagged WT and E196D mutant GSK3α are overexpressed in HEK-293T cells. Western immunoblot for p-GSK3α (Tyr279) and T-GSK3α after FLAG-GSK3α immunoprecipitation and treatment with DMSO or BRD0705 at 10 μM.
